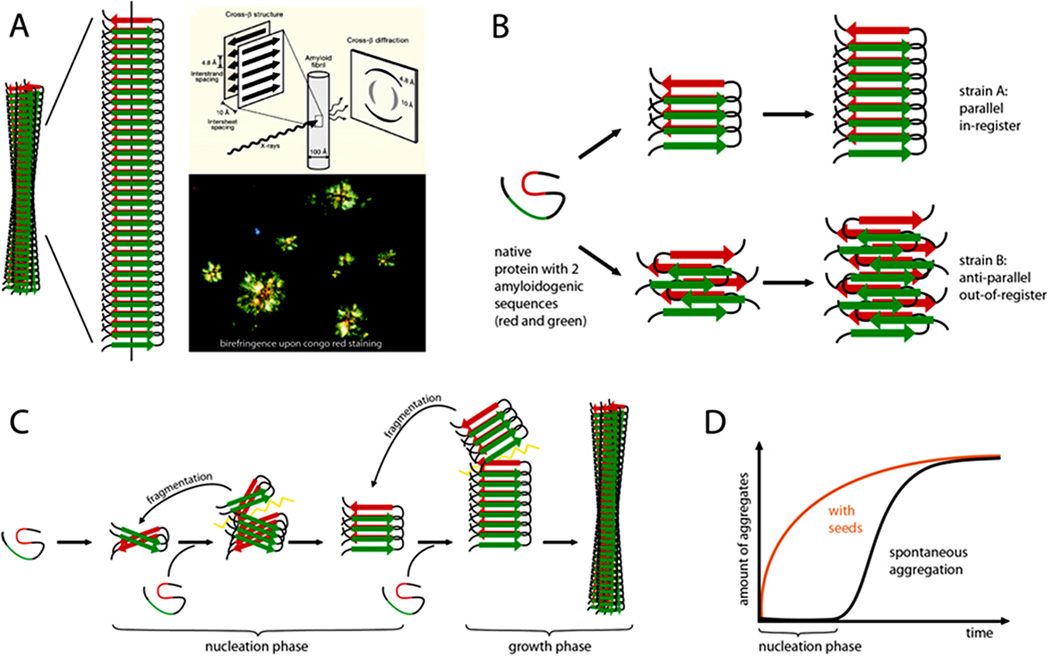
Box 1. The amyloid state of proteins as a framework to explain prion-like protein seeding.
A protein in the amyloid state forms bundles of twisted, unbranched filaments. Each filament is composed of sheets of β-strands (Panel a). These β-sheets run parallel to the filament axis, and the strands are nearly perpendicular to the long axis. This structural arrangement produces a distinctive, cross-β X-ray diffraction pattern that reflects the characteristic spacing between the β-sheets and the β-strands32. Biophysicists classify amyloid based on the X-ray diffraction pattern, whereas pathologists define amyloid as deposits of fibrillar protein in cells or tissues that show reddish/green birefringence under cross-polarized light after staining with the dye Congo red22. Part of panel a is modified, with permission, form ref. 32.
In the most common amyloids, the β-sheets consist of parallel β-strands that are hydrogen-bonded by their backbones. The sheet is ‘in-register’ when identical side chains are on the top of each other. The sheets are bonded to each other via amino acid side chains that are inter-digitated like a zipper. Amyloid ‘steric zippers’ can be formed from identical or different β-strands (homosteric versus heterosteric zippers). An amyloid-forming protein may contribute more than one β-strand segment to the cross-β amyloid backbone (spine)32.
At the molecular level, amyloids can be highly polymorphic; that is, a given β-strand segment is able to form a variety of distinct cross-β amyloid spines and filamentous structures (panel b). Such conformational variants are suggested to be the molecular basis of amyloid ‘strains’, that is, amyloids formed from a particular protein but with different biological activities16,32.
Amyloid formation (panel c) starts with a slow nucleation phase (the aggregation of the protein into a seed) that may go through a series of intermediate states until the initial segment of the amyloid spine is formed21,124. Monomers or oligomeric structures are then bonded to the ends of the initial amyloid seed by conformational conversion. With increasing length, and depending on the conformational stability of the amyloid spine, the growing fibril can eventually break, either spontaneously or actively through cellular processes. In this way, amyloid formation becomes self-propagating through the generation and spread of new amyloid seeds. The kinetics of amyloid fibril formation are a function of the rates of nucleation, growth, and fragmentation21,125. The lag time that precedes protein aggregation in vitro can be greatly shortened by the addition of pre-formed exogenous seeds (panel d).