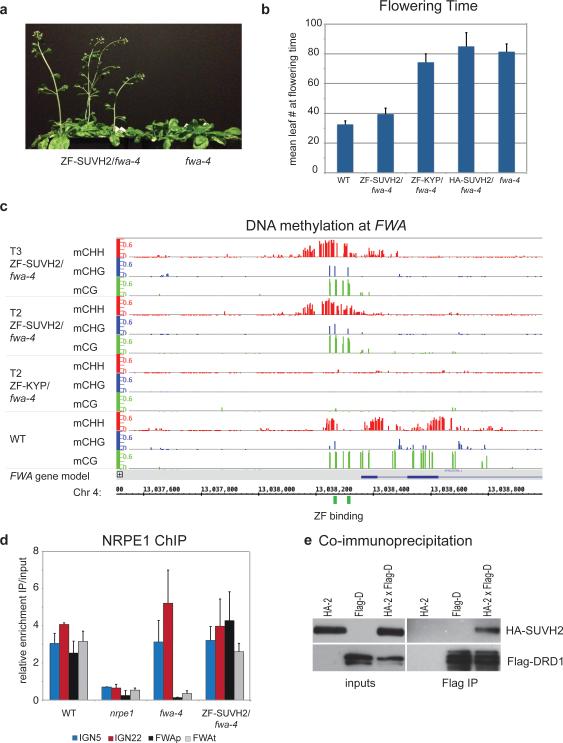
Figure 4. Tethered SUVH2 recruits Pol V through DRD1, resulting in DNA methylation and a late-flowering phenotype.
a. Plants grown side-by-side to illustrate early flowering of ZF-SUVH2 in fwa-4 (T2 plants) compared to fwa-4.
b. Flowering time of Columbia-0 (WT), ZF-SUVH2 in fwa-4, ZF-KYP in fwa-4, HA-SUVH2 in fwa-4, and fwa-4. Flowering time was determined by counting all rosette and cauline leaves up until the terminal flower. The average leaf number and standard deviation of between 20-30 plants was determined. Mean +/− SD.
c. Percent methylation at each cytosine in the FWA repeat region as determined by BS-seq in T2 and T3 ZF-SUVH2/fwa-4 plants compared to T2 ZF-KYP/fwa-4 (unmethylated) and WT (standard methylation pattern). ZF binding sites are shown in green and the FWA gene in blue.
d. NRPE1 ChIP in WT (positive control), nrpe1 mutant (negative control), fwa-4 epiallele, and ZF-SUVH2/fwa-4. qPCR results of two well-characterized NRPE1 binding sites (IGN5 and IGN22) and two regions in FWA (FWAp: promoter; FWAt: transcript) are shown as enrichment of IP/input relative to negative control. Mean +/− SD of two biological replicas.
e. Coimmunoprecipitation of HA-SUVH2 in Arabidopsis using Flag-DRD1. Left panels are inputs from the two parental strains (expressing either HA-SUVH2 (HA-2) or Flag-DRD1 (Flag-D)) and an F2 line expressing both HA-SUVH2 and Flag-DRD1 (HA-2xFlag-d). The right panels show elution from Flag-magnetic beads. Top panels are HA western blots, bottom panels are Flag western blots.