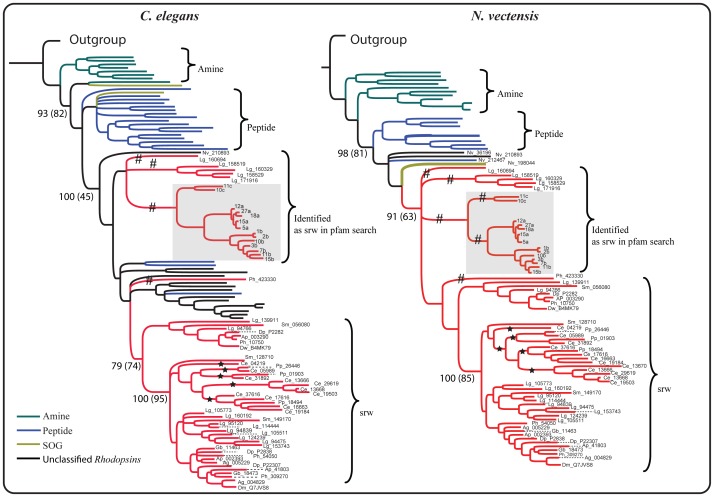
Figure 4. Phylogenetic trees showing closest related Rhodopsin subfamilies to the srw family.
Olfactory like genes identified in N. vectensis is used as outgroup and subsequently rooted. The Rhodopsin subfamily sequences included in the trees were obtained using srw family sequences as queries in BLASTP searches against the Rhodopsin family repertoire of C. elegans and N. vectensis. The top hits included sequences from peptide (violet), SOG (olive green), amine (dark green) subfamilies and some unclassified Rhodopsin family sequences (black) that did not have at least four of the five best hits from the same subfamily in a BLASTP search against human GPCR repertoire (see methods). The topology of trees shows that the peptide (violet) and SOG (olive green) subfamilies are placed basal to the srw clade. Posterior probabilities and the number of bootstrap replicates (within parenthesis) are shown as a percentage for the major nodes. The branches that contain srw family members from nematodes (C. elegans and P. pacificus) are indicated with a star symbol. The branches that contain sequences that are predicted to be srw in the Pfam search and yet separated from the node clustering srw family members from nematodes are indicated with hash (#) symbol. A. californica chemosensory genes that had the highest alignment score against the 7tm_GPCR_srw domain in our Pfam search is highlighted in grey box. See supplementary Figure S1 to view the phylogenetic relationships of the Rhodopsin subfamilies and the srw family in all four analyzed species (C. elegans, N. vectensis, D. melanogaster and T. adhaerens).