Fig. 2.
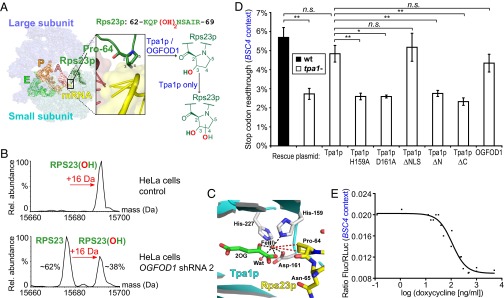
OGFOD1 and Tpa1p catalyze RPS23 trans-3-hydroxyprolyl and dihydroxyprolyl formation, respectively. (A) Schematic depicting Tpa1p/OGFOD1 hydroxylation near the ribosomal decoding center; Rps23p Pro-64 is located at the apex of a loop protruding from a β-strand. View of a structure of the bacterial 70S ribosome including tRNAs and mRNA (yellow), with Rps23p (forest green) modeled onto bacterial S12 [Protein Data Bank (PDB) ID codes 2J00, 2J01, and 3U5C] (17, 30). (B) Deconvoluted ESI-MS spectrum of RPS23 after UPLC-separation of ribosomes from HeLa cells stably transfected with doxycycline-inducible OGFOD1 shRNA 2. Observed masses in control (15,692 Da) and induced (15,676 Da) cells correspond to monohydroxylated and unmodified RPS23 (calculated 15,676 Da; N-terminal methionine cleaved), respectively. (C) View of the Tpa1p active site (cyan/gray; PDB ID code 3KT7) (22) with ribosomal Rps23p (yellow; PDB ID code 3U5C) (30) modeled into the active site (Fig. S3C). Wat, water. (D) Reduced BSC4 stop codon readthrough from TPA1 deletion (P = 0.003) is rescued by a Tpa1p plasmid (P = 0.095). Tpa1p Fe(II) binding (H159A, D161A) and domain deletion (ΔN, ΔC) variants are inactive (P = 0.006, 0.011, 0.008, and 0.004, respectively), whereas Tpa1p ΔNLS (P = 0.54) and OGFOD1 (P = 0.25) are similar to WT Tpa1p. n = 3; mean ± SD; *P < 0.05; **P < 0.01; n.s., not significant. (E) A doxycycline-repressible TPA1 plasmid affects BSC4 stop codon readthrough in a tpa1− strain (IC50 = 110.1 ± 1.2 ng/mL doxycycline; 95% confidence interval 76.1–159.2 ng/mL; R2 = 0.9573; values are pooled from n = 2).
