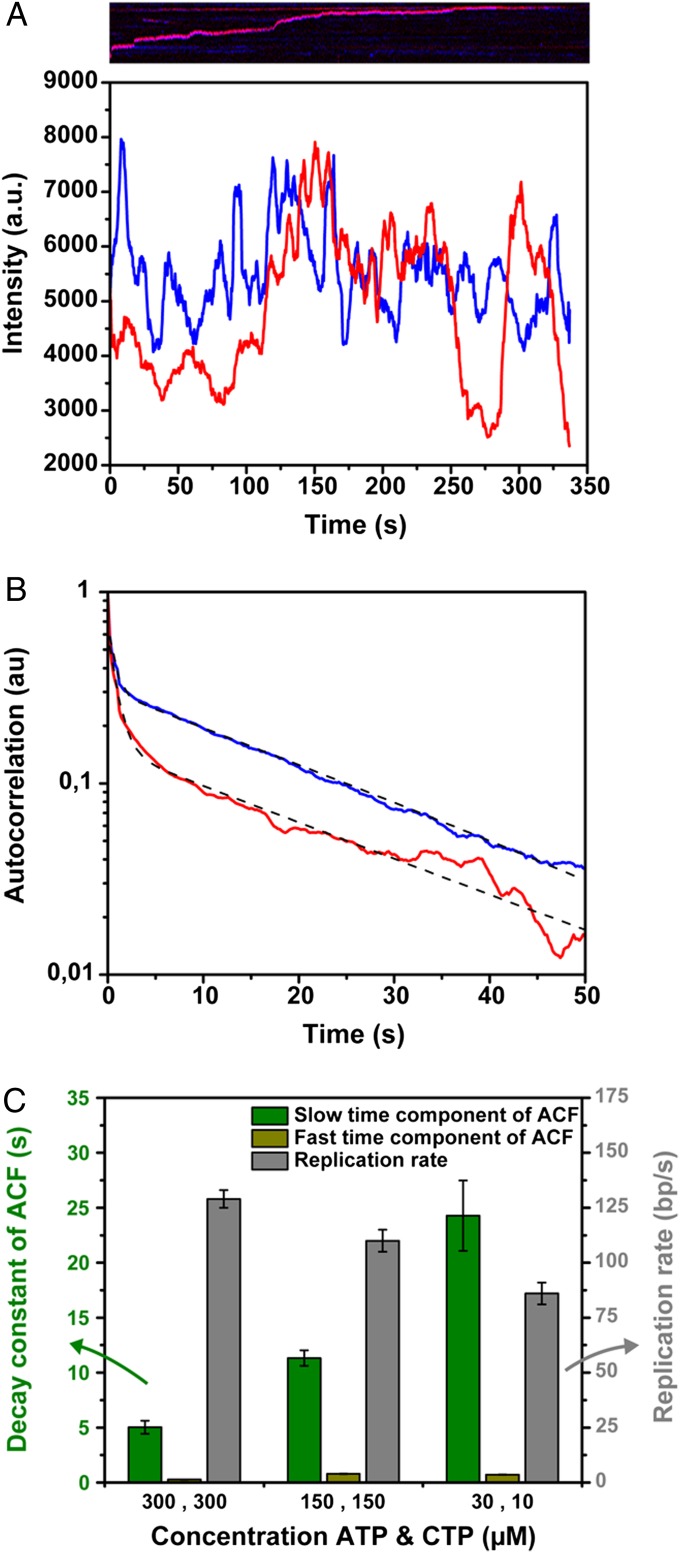
Fig. 2.
Autocorrelation function of the fluorescence intensity of the replisome over time. (A) Fluorescence intensity fluctuations of a single replisome over time. The corresponding kymograph is shown above the graph. The blue line represents the Alexa Fluor 488 intensity, whereas the red line shows the Cy5 intensity at the replisome. (B) Averaged normalized autocorrelation function of the fluorescence intensity of 25 replisomes, replicating DNA in the presence of 150 µM ATP and CTP. The autocorrelation function of the Alexa Fluor 488-labeled polymerases is shown in blue, whereas the red curve represents the autocorrelation function of the Cy5-labeled polymerases. The autocorrelation functions are fitted with a dual exponential decay, where we interpret the fast time scale to represent transient electrostatic binding of gp5 to the carboxy termini of either gp4 or gp2.5 (0.92 ± 0.02 s and 0.66 ± 0.02 s for the red and blue curves, respectively), and the slower time scale shows the replacement kinetics of DNA-synthesizing polymerases at the replisome (22.3 ± 0.5 s for the red and 22.9 ± 0.2 s for the blue curve). (C) Bar plot of the slow and fast time scale of the autocorrelation function and the replication rate as a function of ATP and CTP concentration. The characteristic time scales of DNA-synthesizing polymerase exchange (green bars) are constituted of the slower decay constants of the autocorrelation function divided by 2, to correct for exchange events of polymerases with the same dye attached which decrease the exponential decay of the autocorrelation functions by twofold. The grey bars show the DNA replication rate computed by tracking the position of the replisome over time. Both the polymerase exchange times and DNA replication rates were calculated from trajectories of multiple individual replisomes.