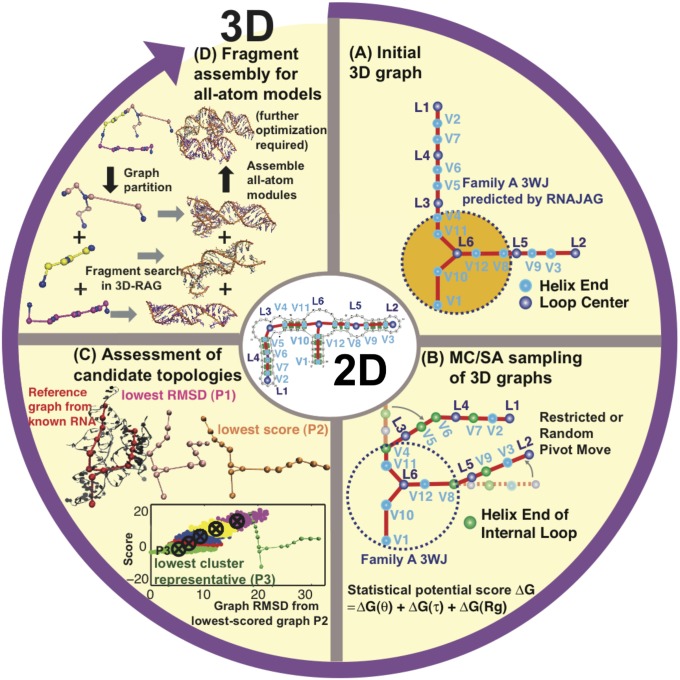
Fig. 1.
Hierarchical MC/SA sampling protocol. (A) Given a 2D structure, an initial planar graph embedded in 3D is constructed by scaling the edges of a 2D tree graph according to size measures. The three-way and four-way junction helical arrangements and coaxial stacking are predicted by RNAJAG (23). (B) From junction geometries and edge lengths, initial planar tree graphs are subject to MC/SA in 3D space guided by knowledge-based statistical potentials for bending and torsion angles of internal loops and radii of gyration. (C) Candidate graphs after MC/SA are selected by lowest rmsd, lowest score, or lowest cluster representatives, and compared with reference graphs translated from solved RNAs. (D) All-atom models are constructed by graph partitioning, fragment search, and assembly of corresponding all-atom modules in 3D-RAG.