Fig. 2.
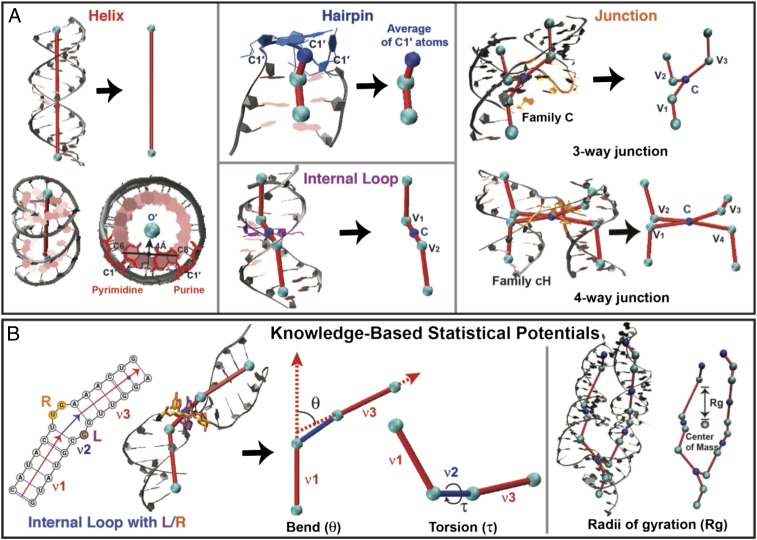
(A) RNA 3D graph representations. Helix ends (cyan) and centers of unpaired regions (hairpins, internal loops, and junctions; blue) are translated into two different classes of vertices. Coordinates for each helix end are defined by the origin of each terminal base pair (O′) (27). Helices are translated to edges. The vertex coordinate representing a hairpin is defined by an average of C1′ atoms of all unpaired bases of a hairpin loop. The Cartesian centroid (C) of an n-way junction is an average of coordinates of n adjacent vertices for n helix ends, as illustrated for internal loops, three-way, and four-way junctions. Edges connect a centroid vertex to adjacent vertices of the proximal helix ends. The three- and four-way junction topologies are predicted by RNAJAG (23). (B) Knowledge-based statistical potentials for bending and torsion of internal loops, and radii of gyration of an RNA 3D graph. An internal loop between double-stranded regions (v1 and v3) connected by a bulge (v2) is defined by L and R bases, where L ≤ R. The bending angle θ is between v1 and v3, and the dihedral angle τ is between v1 and v3 along v2. These angles relate to the size and symmetry of L and R. The radii of gyration measure global compactness by the mean distance from each vertex to the center of mass of all vertices.