Summary
Cell banking, disease modeling, and cell therapy applications have placed increasing demands on hiPSC technology. Specifically, the high-throughput derivation of footprint-free hiPSCs and their expansion in systems that allow scaled production remains technically challenging. Here, we describe a platform for the rapid, parallel generation, selection, and expansion of hiPSCs using small molecule pathway inhibitors in stage-specific media compositions. The platform supported efficient and expedited episomal reprogramming using just OCT4/SOX2/SV40LT combination (0.5%–4.0%, between days 12 and 16) in a completely feeder-free environment. The resulting hiPSCs are transgene-free, readily cultured, and expanded as single cells while maintaining a homogeneous and genomically stable pluripotent population. hiPSCs generated or maintained in the media compositions described exhibit properties associated with the ground state of pluripotency. The simplicity and robustness of the system allow for the high-throughput generation and rapid expansion of a uniform hiPSC product that is applicable to industrial and clinical-grade use.
Graphical Abstract
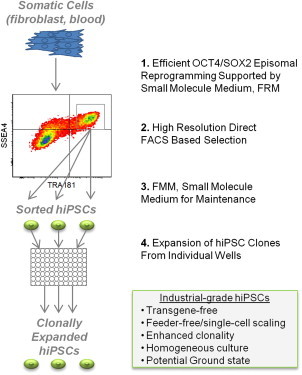
Highlights
-
•
FRM supports highly efficient minimal factor episomal reprogramming
-
•
Clonal derivation of hiPSCs through high-resolution single-cell sorting
-
•
FMM supports long-term maintenance of genomically stable, transgene-free hiPSCs
-
•
Maintenance of homogeneous pluripotent culture resembling the ground state
Cell therapy and banking applications have placed increasing demands on hiPSC technology. Here, Flynn and colleagues demonstrate a simple multiplex selection and maintenance platform utilizing stage-specific small molecule media additives and flow cytometry sorting to derive minimal factor episomal hiPSCs that are genomically stable and resemble ground state pluripotent stem cells. This platform may serve as a path to generate clinically relevant hiPSCs.
Introduction
Today’s human induced pluripotent stem cell (hiPSC)-based disease and toxicology screening efforts and tomorrow’s hiPSC-based auto/allogeneic cell therapies will require robust, reproducible methods of cell line generation and expansion, without the integration of the reprogramming factor transgenes (Saha and Jaenisch, 2009). hiPSCs were first generated by the ectopic expression of multiple genes introduced through genome-integrating retro- and lentiviral expression systems: namely OCT4 (POU5F1), SOX2, cMYC, KLF4, LIN28, and NANOG (Takahashi et al., 2007; Yu et al., 2007). To eliminate as many integrating events as possible, several studies were able to substitute unique small molecule inhibitors for a number of reprogramming factors (Nie et al., 2012). The derivation of footprint-free hiPSCs has been demonstrated using several nonintegrative strategies, including episomal-, RNA-, Sendai virus-, and protein-based methods (Fusaki et al., 2009; Warren et al., 2010; Yu et al., 2009; Zhou et al., 2009). However, nonintegrative methods have proven to be inefficient and labor intensive, often requiring additional reprogramming factors (Lee et al., 2013).
In the most commonly used conventional culture system, human embryonic stem cells (hESCs) and hiPSCs are maintained on feeder cells while passaged as clumps to prevent extensive cell death and genomic aberrations (Thomson et al., 1998). The inability to single-cell culture hiPSCs in a feeder-free (FF) environment severely limits potential industrial scale screening or cell therapy applications (Valamehr et al., 2011). Recent efforts focused on improving single-cell survival have identified small molecule inhibition of the Rho-associated protein kinase to dramatically reduce cell death upon single-cell dissociation (Watanabe et al., 2007; Xu et al., 2010). More recently, we have demonstrated efficient single-cell and FF culture of lentiviral-derived hiPSCs in the presence of a small molecule cocktail, SMC4 (Valamehr et al., 2012). By inhibiting pathways associated with cell death and differentiation, we were able to achieve high-resolution direct flow cytometry sorting and maintenance of hiPSCs in a completely FF system (Abujarour et al., 2013; Valamehr et al., 2012). However, our study focused on lentiviral-derived hiPSCs that were not transgene-free, limiting its therapeutic relevance. Another challenge in pluripotent stem cell culture is their propensity for spontaneous differentiation (Pera and Trounson, 2004; Valamehr et al., 2011). This issue has been resolved for mouse pluripotent stem cells: by blocking differentiation cues of mouse ESCs in culture through small molecule inhibition of mitogen-activated protein kinase and glycogen synthase kinase 3 (termed “2i”), the ground state of pluripotency was achieved, and spontaneous differentiation of mouse ESCs was prevented (Ying et al., 2008). Similar studies in hESCs have been described; however, continuous ectopic expression of pluripotency genes was necessary to maintain the ground state resulting in genome-modified human pluripotent stem cells (Hanna et al., 2010a; Wang et al., 2011).
Here, we describe a multistage culture platform that enables highly efficient episomal reprogramming with a significant reduction in time and effort for hiPSC generation and using just a minimal number of reprogramming factors (OCT4/SOX2/SV40LT). Furthermore, the system can be multiplexed to allow for parallel reprogramming and characterization of multiple somatic cell lines with ease and in an expedited time: bona fide hiPSCs are individually flow cytometry sorted into 96-well plates, characterized, and readily expanded with minimal hands-on effort. We describe improvements in medium composition that allow long-term maintenance of transgene-free hiPSCs in single-cell and FF passage culture with genomic stability and minimal evidence of spontaneous differentiation. Furthermore, gene expression analysis shows that small molecule inhibition of specific signaling pathways may drive hiPSCs to a common ground state of pluripotency, regardless of genetic background and independent of transgene expression. We believe that this demonstration of stable culture of transgene/footprint-free hiPSCs as single cells in a FF environment with the elimination of spontaneous differentiation will serve as a valuable asset in driving pluripotent stem cell technology toward clinical applications.
Results
Identifying Medium Compositions for Long-term Maintenance and Expansion of hiPSCs
We have previously demonstrated improved reprogramming efficiencies and single-cell enzymatic, FF passaging of lentiviral-derived hiPSCs using a small molecule cocktail containing ROCK, GSK3, MEK, and TGF-β pathway inhibitors, termed SMC4 (Valamehr et al., 2012). Although the majority of lentiviral-derived hiPSC lines in SMC4-supplemented cultures maintained a homogeneous population of undifferentiated cells, a subset of lines displayed various degrees of spontaneous differentiation on extended culture (Figure 1A). Analysis of the viral WPRE regulatory element revealed that silencing of the reprogramming factor transgene in the hiPSC lines correlated with the observed spontaneous differentiation (Figure 1B). We therefore initiated an assessment of the cell culture components, including the small molecule inhibitors, in order to identify whether a homogeneous pluripotent population could be maintained during continuous culture on FF and single-cell enzymatic passage regardless of residual transgene expression. We observed that inhibition of the TGF-β pathway during long-term maintenance was the most significant factor in inducing the spontaneous differentiation of hiPSC lines with silenced transgene expression (Figure 1C). This observation was consistent with previous studies demonstrating that the TGF-β pathway plays opposing roles in reprogramming and maintenance (Beattie et al., 2005; Lin et al., 2009). We therefore derived a medium formulation: Fate maintenance medium (FMM) containing ROCK, GSK3, and MEK pathway inhibitors, basic fibroblast growth factor (bFGF), and leukemia inhibitory factor (LIF) (Table S1 available online). When hiPSC clone FTi096 was directly transitioned to FMM, the majority of the spontaneous differentiation was eliminated within two to three passages, without the need for additional culture manipulation, resulting in a homogeneous population of SSEA4/TRA181-positive cells (Figure 1A).
Figure 1.
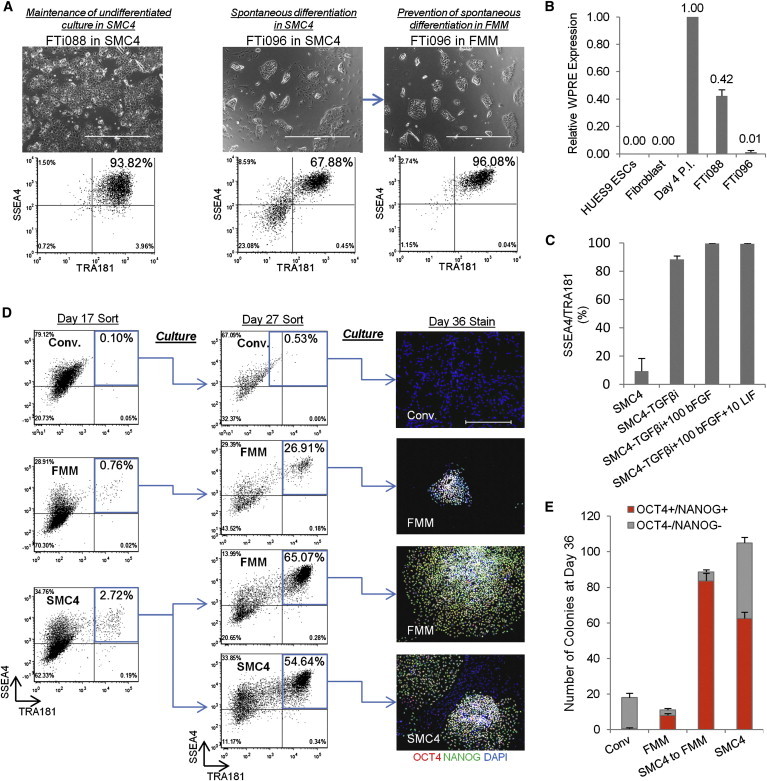
Multistage Medium Platform for Enhanced Reprogramming and hiPSC Maintenance
(A) Lentiviral-generated hiPSC clones FTi088 and FTi096 cultured in SMC4. The spontaneous differentiation of clone FTi096 is minimized when transitioned to FMM for three passages shown by morphology (upper panels) and flow cytometry profile (lower panels). Scale bars, 1,000 μm.
(B) Quantitative real-time PCR for transgene expression of viral element WPRE. Expression was normalized to GAPDH and relative to WPRE expression of parental fibroblast line 4 days post-lentiviral infection (Day 4 P.I.). Three independent experiments were performed with an SEM.
(C) SSEA4 and TRA181 status of transgene-silenced lenti-induced hiPSCs in various components: removal of SB431542 (−TGF-βi), 100 ng/ml bFGF (+100 bFGF), and 10 ng/ml LIF (+10 LIF). Three independent experiments were performed with an SEM.
(D) Fibroblast line was transfected with lentiviral construct containing gene set OKS, cultured, and sorted as indicated. Sort gate is highlighted in blue. At day 27, the cultures were resorted, seeded at normalized density, and maintained in respective media for an additional 9 days. Panels on the right are representative colonies. Scale bars, 500 μm.
(E) Colony counts of day 36 staining as discussed in (D): p < 0.0005 between SMC4 versus SMC4 to FMM OCT4−/NANOG− colony number. Two independent experiments were performed for conventional (Conv.) and FMM, and three independent experiments were conducted for SMC4 to FMM and SMC4, with an SEM.
See also Table S1.
To test whether FMM can completely replace SMC4-supplemented medium during the reprogramming process, we initiated OCT4/KLF4/SOX2 (OKS) lentiviral reprogramming in conventional culture (hESC medium on mouse embryonic fibroblast [MEF] feeder cells), SMC4-supplemented medium in FF culture, or FMM in FF culture (Figure 1D). Seventeen days after the induction of lentiviral reprogramming, the cells from each culture were flow cytometry sorted, and cells positive for SSEA4/TRA181 were selected and replated in their respective culture media. The reprogramming cells from the SMC4 culture were replated in either SMC4 or FMM for comparison (Figure 1D). As previously seen, SMC4 dramatically improved the kinetics of reprogramming, resulting in significantly more SSEA4/TRA181-positive cells at day 17 postinduction (2.72% versus 0.76% for FMM and 0.10% for conventional culture; Figure 1D) (Valamehr et al., 2012). The cultures were maintained for a total of 36 days postinfection and scored for undifferentiated colonies (Figures 1D and 1E). The combination of initial reprogramming in SMC4 followed by a transition to FMM ultimately resulted in more OCT4/NANOG-positive colonies and a significantly reduced number of OCT4/NANOG-negative colonies relative to continuous maintenance in SMC4 (Figures 1D and 1E). In cultures maintained exclusively in FMM, the number and size of the colonies appeared inferior to the stage-specific media approach. Cultures continuously maintained in conventional hESC media on feeder cells did not produce any OCT4/NANOG colonies, consistent with the low efficiencies seen by OKS reprogramming when unaided by small molecules (Nakagawa et al., 2008). Therefore, a multistage culture system that targets unique pathways at different stages of the reprogramming and maintenance process appears to be the most efficient and robust approach to hiPSC generation.
Multiplex Generation, Characterization, and Maintenance of Transgene-free hiPSCs in a Single-Cell Passage and FF Format
Although attractive for clinical use, episomal vector systems for nonintegrative reprogramming are extremely inefficient (<0.001%), especially in FF environments (Narsinh et al., 2011; O’Doherty et al., 2013). We therefore assessed whether episomal induction would be best supported by a multistage culture system. We adapted our hiPSC generation platform as outlined in Figure 2A: 24 hr after induction of episomal expression, the reprogramming culture is transitioned to Fate-reprogramming media (FRM) to enhance reprogramming kinetics, and at day 14, the culture is switched to FMM to promote hiPSC proliferation in an undifferentiated state. Between days 16 and 22, the culture is flow cytometry analyzed for individual hiPSCs using a cocktail of surface markers geared for identification of bona fide hiPSCs (Abujarour et al., 2013) and directly sorted into FF 96-well plates at clonal densities, i.e., three events per well. The 96-well plates are then surveyed by various criteria including expression of key pluripotency markers, and selected hiPSC clones are expanded as single cells in a FF environment with FMM (Figure 2A).
Figure 2.
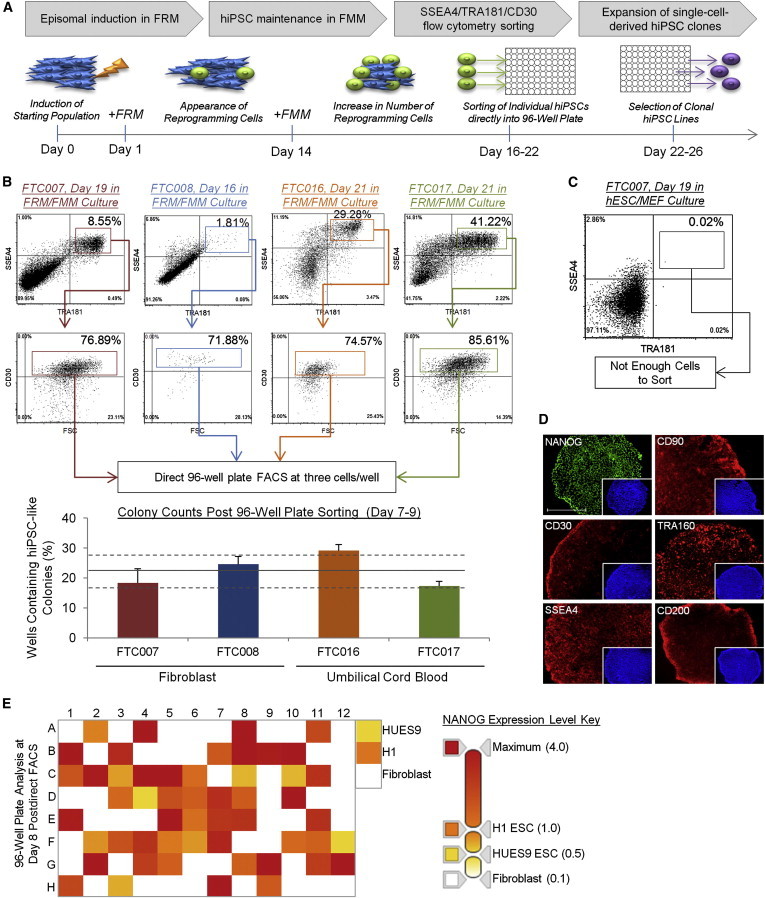
Individual Episomal-Reprogrammed hiPSCs Are Efficiently Selected and Seeded in 96-Well Plates for Clonal Expansion
(A) Schematic timing illustration of episomal induction, multistage media system, flow cytometry sorting, and clonal expansion.
(B) Flow cytometry profile of episomal-induced reprogramming maintained in FRM-to-FMM transition in FF culture (outlined in A) at indicated days post-transfection. Sort-gating strategy used for each parental line (SSEA4+/TRA181+/CD30+ population) is illustrated in respective colors, corresponding to the bottom histogram panel representing the percentage of wells of 96-well plate containing individual hiPSC clones. Wells containing multiple clones or differentiated clones were not scored. Three independent experiments were performed with an SEM. The solid line represents the average percentage among all derivations with dotted lines representing SD, 22.3 ± 5.5.
(C) Flow profile of FTC007 induced to reprogram 19 days posttransfection maintained in conventional medium in the presence of MEF cells. The induced population is taken from the same population of FTC007 in (B), however, treated in different culture thereafter.
(D) Immunocytochemistry analysis of various pluripotency markers of sorted colonies in 96-well plate. Right corner panels represent DAPI staining. Scale bar, 500 μm.
(E) Quantitative real-time PCR for NANOG expression for each well of a SSEA4/TRA181/CD30 direct sorted (FACS) 96-well plate at three cells per well. The expression range is between zero and four times expression relative to H1 human ESCs as described in the legend located on the right side and normalized to GAPDH.
See also Figures S1, S2, and S3.
In a pilot study, an episomal expression system consisting of gene combination OCT4/SOX2/NANOG/KLF4/LIN28/MYC/SV40LT (OSNKLMT) (Yu et al., 2009) was used to transfect fibroblast cells. By day 10, a relatively large population of SSEA4/TRA181-positive cells was detected (>1.0%, Figures S1A and S1B). This compared favorably to the efficiency seen in other episomal-reprogramming systems (Narsinh et al., 2011). We also noticed that the reprogramming colonies were extremely heterogeneous, placing further value in the ability to sort and select individual cells compared to traditional manual colony picking (Figure S1B). On day 14, the reprogramming culture supported by FRM was split into either FRM or FMM. On day 21, each culture was flow cytometry sorted for SSEA4/TRA181/CD30-positive cells (Figure S1C). Cultures maintained in FRM contained a mixed population of differentiated and undifferentiated cells, whereas the culture switched to FMM contained mostly undifferentiated cells (Figure S1D).
In order to explore the throughput and robustness of this approach, we applied it to different somatic parental lines sourced as either fibroblasts or CD34+ cells expanded from mobilized peripheral blood or minimal volumes of umbilical cord blood from donors of different ages, genders, and ethnicities (Figures S2A and S2B). Each line was induced to reprogram as outlined in Figure 2A with the episomal gene combination set OSNKLMT and 96-well plate flow cytometry sorted for individual hiPSCs (Figures 2B, S2C, and S2D). A large population of SSEA4/TRA181/CD30-positive cells was observed for the majority of our reprogramming lines, and the effectiveness of the FRM and FMM system was further highlighted when compared to a parallel reprogramming experiment using conventional medium and feeder cells (8.55% in FRM/FMM versus 0.02% in conventional culture for the FTC007 fibroblast line; Figures 2B and 2C). On average, 22 clonal hiPSCs per 96-well plate were seen for each somatic line (Figures 2B, S3A, and S3B). Colonies were subsequently confirmed as bona fide hiPSC clones by direct analysis of intracellular and surface marker expression and direct quantitative real-time PCR for NANOG (Figures 2D, 2E, and S3C). A similar reprogramming efficiency was observed with the defined surface coating Vitronectin (Figure S3D). This data set demonstrates that the platform is robust and reproducible when applied to episomal reprogramming, including in single-cell and FF culture, and allows for multiple reprogramming experiments to be performed in parallel with minimal effort and without compromising quality.
Long-Term Passage and Expansion of Transgene-free hiPSC Lines in FMM
In order to assess the long-term passage and expansion of hiPSCs in our FMM platform, clones selected from the reprogramming study were expanded as single cells in FF culture (Figures 3A and S3E). hiPSC lines seen to have lost episomal DNA by passage 4–7 were carried forward for analysis (Figure 3B). Quantitative real-time PCR analysis of pluripotency markers confirmed the undifferentiated state of selected hiPSC lines, with KLF4 expression being noticeably elevated (Figure 3C). Immunofluorescence expression and flow cytometry analyses also showed hiPSC lines to maintain a homogeneous population of undifferentiated cells (Figures 3D and 3E). The homogeneity of culture was maintained without the need of any cleanup strategies that are commonly utilized in pluripotent culture, such as the manual separation of differentiated from undifferentiated colonies. Similar expansion of uniform hiPSC cultures was observed when Matrigel was replaced with the defined surface coating Vitronectin during routine single-cell passaged culture (Figures S3F–S3H).
Figure 3.
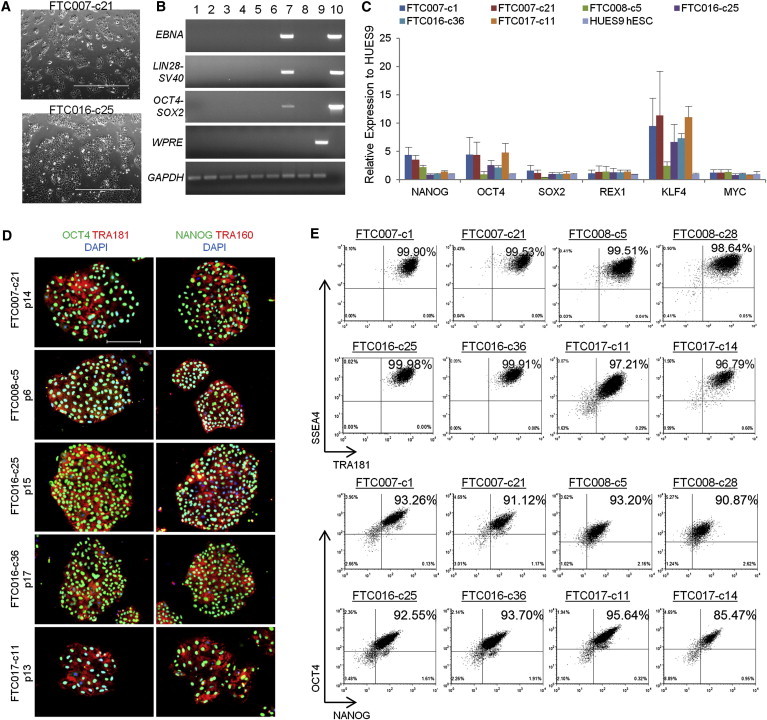
Episomal-Reprogrammed hiPSC Clones Maintain Their Undifferentiated State and Are Free of Transgene Sequence
(A) Representative images of hiPSC clone during culture. Scale bars, 1,000 μm.
(B) PCR analysis for episomal DNA derived from various hiPSC clones. Lane 1 shows FTC007-c1 p4, lane 2 shows FTC007-c21 p4, lane 3 shows FTC016-c25 p5, lane 4 shows FTC016-c36 p5, lane 5 shows FTC017-c11 p7, lane 6 shows FTC017-c14 p7, lane 7 shows FTC017-c17 p6 (a line maintaining episomal constructs used a positive control), lane 8 shows untransfected FTC007, lane 9 shows hiPSC generated using lentiviral constructs, and lane 10 shows episomal vector used as positive control. Input of 100 ng genomic DNA and 35 PCR cycles were used for all sets.
(C) Quantitative real-time PCR analysis for endogenous pluripotent gene expression. Data were normalized to GAPDH and relative to HUES9 hESCs. Four independent experiments were performed, with an SEM.
(D) Pluripotency markers detected by immunofluorescence. Scale bar, 200 μm.
(E) Flow cytometry profile for selected hiPSC clones from various parental lines.
See also Figure S3.
Genomic abnormalities are often detected in hESC and hiPSC lines passaged as single cells in a FF environment (Laurent et al., 2011; Taapken et al., 2011). Importantly, hiPSC clones single-cell and FF cultured in FMM for an extended period (25–30 passages) continued to maintain their undifferentiated profile and genomic stability without the need for culture cleaning or further selection (Figures 4A and 4B). When induced to differentiate, episomal-derived hiPSC clones maintained in FMM readily gave rise to all three somatic lineages (Figures 4C–4E).
Figure 4.
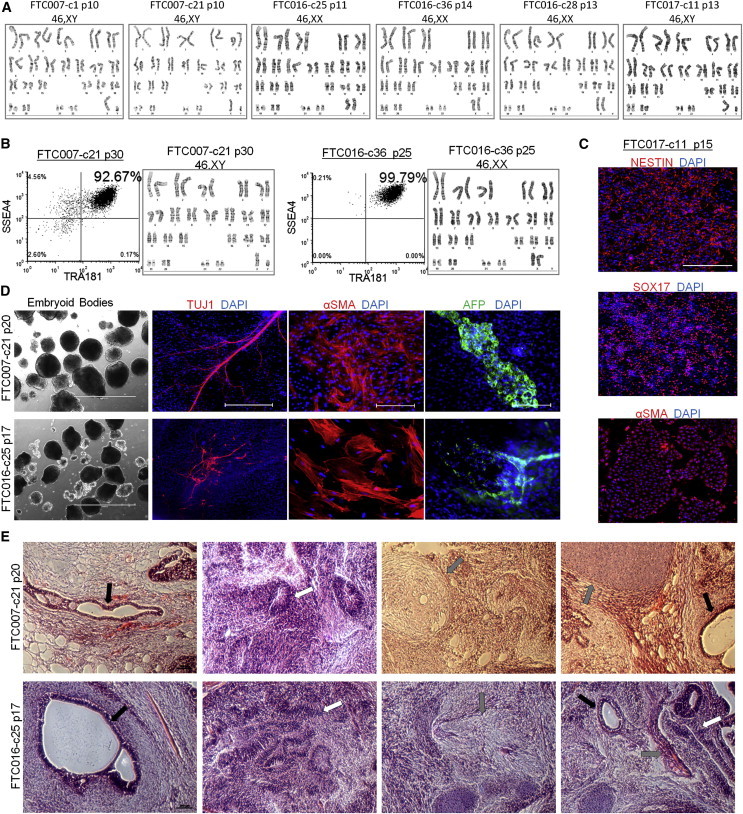
Genomic Stability and Pluripotency Are Maintained during Continuous Single-Cell and FF Culture
(A) Cytogenetic analysis on 20–40 G-banded metaphase cells from various hiPSC clones maintained in FF and single-cell culture.
(B) Flow cytometry profile and cytogenetic analysis of long-term-passaged hiPSC clones in FF and single-cell enzymatic-passaged culture.
(C) Three to 4-day-directed differentiation of FTC017-c11. Scale bar, 200 μm.
(D) Embryoid body formation and differentiation. Immunocytochemistry was conducted 28 days post-differentiation: ectoderm, Tuj1; mesoderm, α smooth muscle actin (aSMA); and endoderm, AFP. Scale bars, 1,000 μm (bright-field images), 200 μm (TUJ1 images), and 500 μm (SMA and AFP images).
(E) Histological sections of teratoma derived from FTC007-c21 and FTC016-c25. Black arrows point to endoderm, white arrows point to ectoderm, and gray arrows point to mesoderm. Scale bar, 200 μm.
Enabling Episomal Reprogramming with Minimal Genes
Having demonstrated an extremely efficient and robust platform for the generation and expansion of transgene-free hiPSCs using OSNKLMT-reprogramming factors, we next wanted to test if this same system could enable hiPSC generation without the requirement for many reprogramming factor genes. An efficient footprint-free expression system with the reduced dependency for oncogenes such as KLF4, MYC, and LIN28 or the need to knock down P53 during the reprogramming process would be of great value for pluripotent stem cell therapies (Lee et al., 2013; Okita et al., 2011; Yu et al., 2009). We therefore constructed several episomal expression cassettes lacking the oncogenic-reprogramming factors and containing minimal gene sets including OCT4/NANOG/SOX2 (ONS), OCT4/SOX2 (OS), or OCT4/OCT4 (2xO) in an attempt to vary gene expression combination and dosage (Figure S4A).
Reprogramming of fibroblast line FTC007 with various combinations of OCT4, NANOG, SOX2, and SV40LT was attempted in the FRM, FMM, and flow cytometry selection platform as previously described (Figure 2A). Although SV40LT alone did not produce any true SSEA4/TRA181-positive cells at day 13 of reprogramming, it was seen to be necessary for improved cell survival post-transfection (Figures 5A and S4B). The various reprogramming factor combinations, including just OCT4/SOX2/SV40LT, resulted in efficient reprogramming as demonstrated by the emergence of SSEA4/TRA181-positive populations early in the reprogramming process (Figure 5A). Surprisingly, the percentages observed were on par with lentiviral and episomal-induced systems containing KLF4 and MYC (Figures 2B, 5A, and S4C). Several reprogramming factor combinations were carried forward and transitioned to FMM prior to flow cytometry sorting and selection of individual hiPSC clones. Similar to OSNKLMT episomal reprogramming, clonal hiPSC lines were readily derived from combinations containing minimal genes OCT4, SOX2, and SV40LT as well as other combinations (Figure 5B). The hiPSC clones that were demonstrated to be negative for transgenes and episomal vector markers by passages 5–7 were carried forward for further analysis (Figure 5C). To provide further evidence for the absence of episomal DNA in hiPSC lines, we conducted a functional test for survival in the presence of hygromycin. Various FCT007-derived hiPSC lines (seen to be karyotype normal while maintaining pluripotency, Figures 2, 3, 4, and 5) were treated with hygromycin at or above the kill concentration (50 and 100 μM, respectively, Figures S4D–S4F). Although survival was seen at both concentrations for the reprogramming pool, none of the tested hiPSC lines demonstrated resistance to hygromycin, indicative of complete loss of episomal DNA (Figures S4E and S4F).
Figure 5.
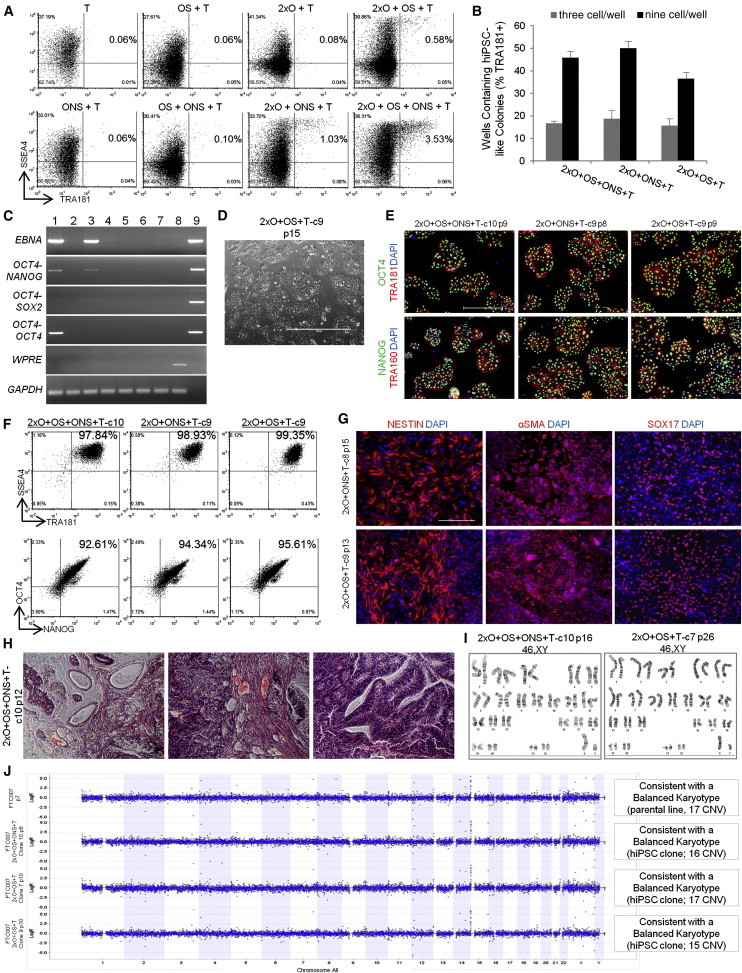
Derivation of hiPSC Clones with Minimal Number of Reprogramming Factors
(A) Flow cytometry profile of reprogramming kinetics induced by various gene combinations at day 13 post-induction.
(B) Histogram of hiPSC clones in wells of 96-well plate at three and nine cells per well. Three independent experiments were performed, with an SEM.
(C) PCR analysis for episomal DNA derived from various hiPSC clones. Lane 1 shows 2xO+OS+ONS+T-c7 p6, lane 2 shows 2xO+OS+ONS+T-c10 p6, lane 3 shows 2xO+ONS+T-c5 p5, lane 4 shows 2xO+ONS+T-c9 p5, lane 5 shows 2xO+OS+T-c7 p6, lane 6 shows 2xO+OS+T-c9 p6, lane 7 shows untransfected FTC007, lane 8 shows hiPSC generated using lentiviral constructs, and lane 9 shows episomal vector used as positive control. Input of 100 ng genomic DNA and 35 PCR cycles were used for all sets.
(D) Typical morphology. Scale bar, 1,000 μm.
(E) Pluripotency markers detected by immunofluorescence. Scale bar, 1,000 μm.
(F) Flow profile of hiPSC clones.
(G) Directed differentiation of selected hiPSC clones 72–96 hr post-induction. Scale bar, 500 μm.
(H) Histological sections of teratoma derived from hiPSC clone 2xO+OS+ONS+T-c10. Left panel shows endoderm, middle panel shows mesoderm, and right panel shows ectoderm. Scale bar, 200 μm.
(I) Cytogenetic analysis of 20 G-banded metaphase cells from various hiPSC clones maintained in FF and single-cell culture.
(J) Copy number variation as assessed by array-comparative genomic hybridization and SNP. Panel on the right represents interpretation summary of the data. CNV, copy number variation.
See also Figure S4.
Selected clones were continuously passaged as single cells in a FF environment and were demonstrated to maintain a homogeneous population of undifferentiated cells, while displaying the ability to efficiently differentiate into the three somatic lineages (Figures 5E–5H). Karyotype and copy number variation analysis revealed genomically stable hiPSC lines during long-term culture maintained in FMM (Figures 5I and 5J). Collectively, the data show that hiPSCs can be readily generated by transient expression of minimal reprogramming genes from episomal systems in the FRM/FMM, flow cytometry-based-reprogramming platform, including a combination consisting of only OCT4, SOX2, and SV40LT.
FMM Supports hiPSC Properties Resembling the Ground State of Pluripotency
We next investigated the gene expression differences between small molecule and conventionally maintained hiPSC cultures (Hanna et al., 2010b; Saha and Jaenisch, 2009). In an initial study, a lentiviral-induced hiPSC clone FTi111 generated and maintained in small molecule culture and shown to be pluripotent was thawed directly into various culture environments (Figures S5A–S5C). During three passages, each culture set demonstrated unique colony morphology (Figure S5D). Interestingly, gene expression of pluripotent markers revealed a distinct pattern between the culture environments (Figure S5E). Although derived from the same thaw, the conventionally maintained culture on feeder cells better resembled hESC control H1 maintained in conventional culture than its counterpart cultures maintained in small molecules (Figure S5E). Both small molecule cultured sets appeared to have elevated gene expression of certain pluripotency genes, including KLF4, whereas other genes such as LIN28 and DPPA4 appeared to be similar between the culture systems (Figure S5E). The differentiation marker, PAX7, appeared to be nearly silenced when hiPSCs were cultured with small molecules in FF culture (Figure S5E).
Continuing these observations, differences between hiPSCs derived using lentiviral induction and conventional hESC/feeder culture and episomal lines derived with different combinations of reprogramming factors maintained in the FRM/FMM platform were assessed. High-content quantitative real-time PCR analysis was used to explore gene expression associated with pluripotency and differentiation. The majority of the pluripotency genes surveyed displayed a similar expression pattern between hiPSCs maintained in FMM culture or conventional cultures containing feeder cells regardless of the reprogramming method or gene combinations used in their derivation; with the exception of KLF4 expression, again seen to be elevated in small molecule-mediated culture (Figure 6A). However, expression differences between cell lines were clearly observed in genes associated with differentiation (Figure 6B). Regardless of the gene combination used during induction, FMM-maintained hiPSCs displayed lower expression of most genes associated with the three somatic lineages when compared to both hiPSCs and H1 hESCs maintained in conventional medium and on feeder cells. A subset of lines induced by episomal gene set OSNKLMT appeared to show expression of ectoderm lineage, OTX2 and TUJ1, whereas this expression was negligible in the hiPSCs episomally derived without the use of Lin28, KLF4, and c-MYC (Figure 6B). The expression of all differentiation genes tested was fully suppressed in all hiPSCs derived from episomal minimal gene sets and maintained in FMM (Figure 6B).
Figure 6.
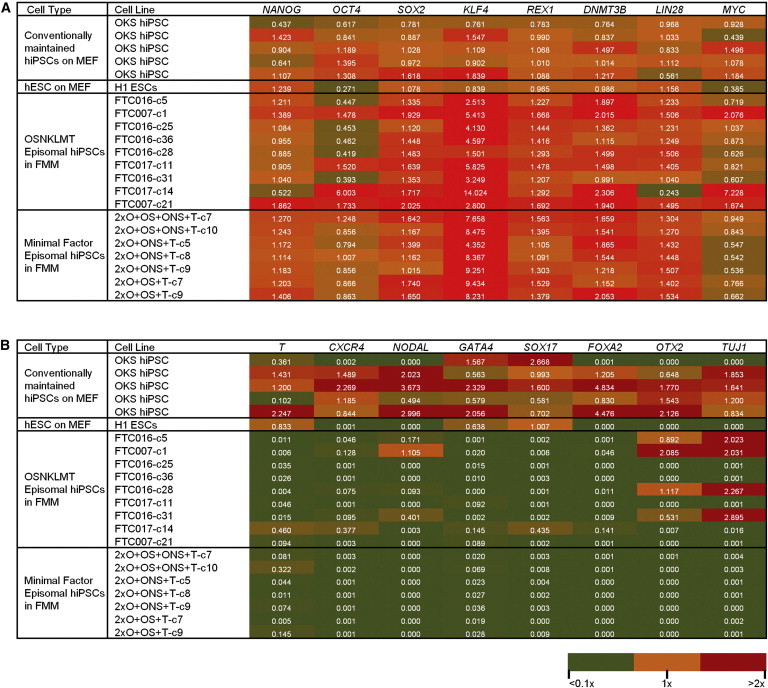
Relative Gene Expression Profile of Minimal Factor Episomal-Induced hiPSCs in FMM
(A and B) Heatmap results derived from a Fluidigm dynamic array depicting relative gene expression levels of pluripotency (A) and differentiation (B) genes of conventionally maintained hiPSC lines, RNA from conventionally maintained H1 hESCs, and episomal hiPSC lines derived using various gene combinations maintained in FMM. Relative gene expression for each line is noted within each box and color coded based on three expression levels summarized in the legend (lower right). All sets were normalized to the average expression of two housekeeping genes (GAPDH and HPRT1) and referenced to the median expression level of six control conventional lines (OSK hiPSCs and H1 hESCs on MEF) representing 1× value for each gene. See also Figure S5.
We next assessed whether hiPSCs derived and maintained in from various media could be defined by patterns in global gene expression analyses. We compared (1) episomal induction maintained in FMM, (2) episomal induction maintained in FMM but switched to conventional medium for three passages, (3) lentiviral induction maintained in SMC4, and (4) lentiviral induction maintained in conventional culture (Figures S6A and S6B). All lines had previously been demonstrated to be pluripotent, genomically stable, and able to differentiate to all three somatic lineages. Cluster analysis of genes differentially expressed between small molecule culture and conventional culture revealed that the hiPSC lines grouped based on current culture conditions, and not by original derivation method and culture (Figure S6B). Gene ontology classification of 300 genes displaying 2.5-fold expression differences identified differentiation and development as the main categories highly enriched in the conventional culture group, whereas genes upregulated in the small molecule culture group were mostly associated with regulation of cell proliferation and sex development (Figures S6C and S6D). Gene expression analysis was repeated with direct comparison of cultures in FMM, conventional, or transitions from each culture to the other (FMM, Conv, FMM→Conv, Conv→FMM; Figure S6A). Again, cluster analysis produced two groups separated based on the current culture system regardless of method of generation or the prior culture system (Figure 7A). For example, FTC016-c28 (FTC016-derived hiPSC clone 28) was generated and maintained under the FRM/FMM platform, prior to transition to conventional culture (Epi-FTC016-c28: FMM→Conv). When analyzed, it grouped with cultures maintained exclusively in conventional culture, and not with its parental line maintained in FMM. The inverse was also seen: lines generated in conventional culture grouped within the conventional set until transition to FMM, upon which it grouped within the FMM cluster (Figure 7A). Gene ontology once again categorized conventional culture to be enriched with genes associated with differentiation and development (i.e., p = 2.2 × 10−10, pattern specification process; Figures 7B and S6E). These results suggest that genes associated with differentiation propensity are reduced in FMM culture, and hiPSCs can be adapted to this system to reduce spontaneous differentiation potential.
Figure 7.
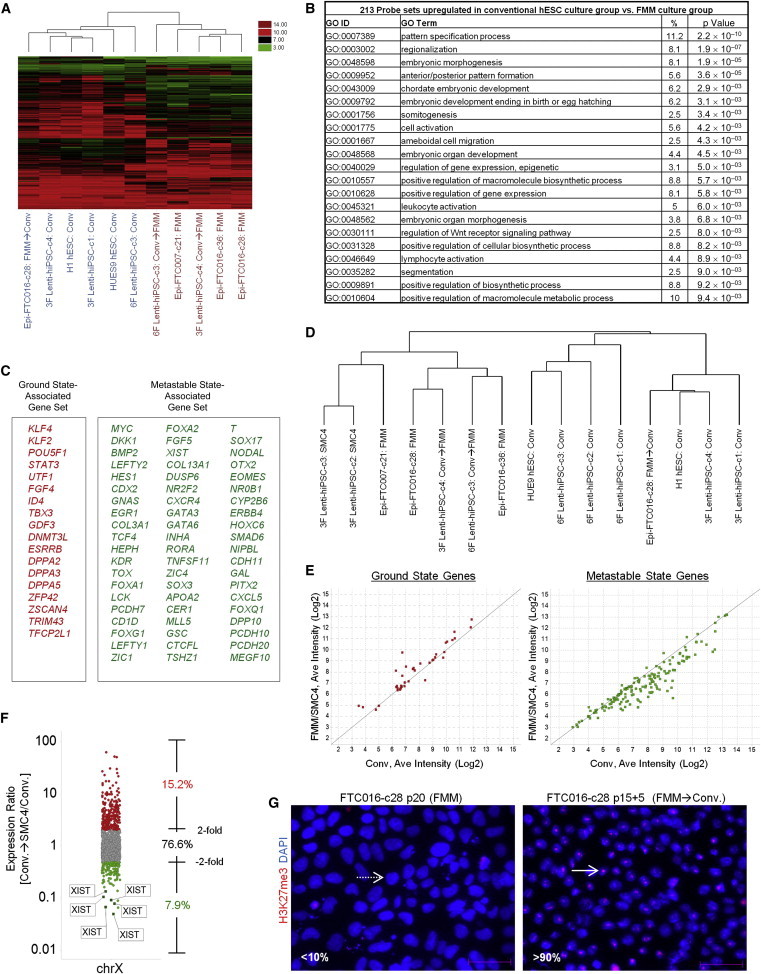
FMM-Maintained hiPSCs Have Reduced Expression of Differentiated Genes and Increased Expression of Genes Associated with the Ground State
(A) A total of 339 probe sets were differentially expressed between conventional and FMM culture by greater or less than 2.5-fold. Hierarchical clustering on the 339 probe sets using a complete linkage method based on Euclidean distance measurements was performed. Conventional culture is noted in blue, and FMM culture is noted in brown.
(B) Gene ontology biological process enrichment analysis (D.A.V.I.D.) of the 213 probe sets upregulated 2.5-fold or greater with conventional culture (in comparison to FMM culture).
(C) Gene lists representative of ground or metastable pluripotency states. List is derived from references noted in text.
(D) Hierarchical clustering on the 227 probe sets corresponding to the genes in (C) using a complete linkage method based on Euclidean distance measurements.
(E) RMA (log2)-normalized intensities for the probe sets corresponding to the genes in (C). Left panel represents 39 probe sets for ground state; right panel represents 188 probe sets for metastable state. Average conventional culture intensity levels are plotted on the x axis, whereas the average FMM/SMC4 intensity is on the y axis, and black line indicates equal expression.
(F) Gene expression comparison of X chromosome-located genes between hiPSC clone derived and cultured in conventional medium culture and its counterpart adapted to SMC4 culture using all 1,688 Affymetrix probe sets mapped to the X chromosome. Probe sets associated with XIST gene expression are highlighted.
(G) Representative images of HEK27me3 on hiPSC clone maintained in FMM or adapted to conventional culture for five passages. Dotted arrow in the left panel points to a representative nucleus absent of H3K27me3 staining, whereas solid arrow in the right panel points to a nucleus positive for H3K27me3 staining. Percentages of nucleus-positive staining are indicated in the lower-left side of each panel. Scale bar, 50 μm.
See also Figures S5 and S6.
With the demonstrated high survival rates in single-cell dissociation and significant suppression of differentiation-related gene expression, we examined if FMM was supporting the ground state of human pluripotent stem cells by blocking pathways associated with differentiation (Ying et al., 2008). Gene lists were compiled to represent ground and metastable states of human pluripotent stem cells (De Los Angeles et al., 2012; Han et al., 2011; Hanna et al., 2010a; Hirata et al., 2012; Nichols and Smith, 2012; Valamehr et al., 2012; Zhou et al., 2010) (Figure 7C). Gene clustering based on these gene lists was then performed for hiPSC lines in FMM or conventional culture (Figure 7D). Similar to the global gene expression comparison, the focused gene clustering showed a clear separation of the cell lines based on their current culture conditions with profiles appearing to be interconvertible. For example, FTC016-c28 transitioned from FMM to conventional culture grouped with H1 hESCs and not with its parental hiPSC line maintained in FMM (Figure 7D). Similarly, a lentiviral hiPSC clone derived from a fibroblast line (6F Lenti-hiPSC-c3) maintained in conventional culture grouped with HUES9 hESCs and other hiPSC clones in conventional culture; however, when switched to FMM, it grouped with an episomal hiPSC derived from umbilical cord blood as well as other FMM-cultured lines (Figure 7D). To determine the distribution of genes representative of the ground and metastable states within the two clusters, we plotted average intensities for each probe set with respect to small molecule (FMM/SMC4) versus conventional culture (Figure 7E). Interestingly, the majority of genes associated with the ground state were seen to have elevated expression in small molecule culture cluster, whereas an increase in expression of genes associated with metastable state was detected in the conventional culture cluster (Figure 7E).
We next analyzed the X-inactivation state of female hiPSCs derived and maintained in conventional culture conditions compared to its counterpart adapted to small molecule culture and maintained for ten passages (Figure 7F). hiPSCs maintained in small molecule culture revealed a notable increase in X chromosome gene expression when compared to conventional culture, suggestive of reactivation of the silenced X chromosome (Figure 7F). The noticeable exception was the X-inactive-specific transcript (XIST), the major effector of X inactivation, which was downregulated in the switch to small molecule culture (Figure 7F). Further evidence of X activation was provided by the differential staining of H3K27me3 in hiPSCs cultured in FMM relative to their counterpart culture adapted to conventional medium (<10% H3K27me3 staining in FMM compared to >90% H3K27me3 staining in conventional culture; Figure 7G). Collectively, these data suggest that FMM represses differentiation cues and in turn promotes features associated with the ground state of pluripotency.
Discussion
Here, we describe a multistage process for highly efficient nonintegrative cellular reprogramming coupled to a method of scalable hiPSC culture that utilizes stage-specific media formulations: FRM for enhanced reprogramming, and FMM for long-term maintenance. Previous studies have demonstrated the versatility of small molecules in the reprogramming and maintenance of pluripotent stem cells of various species (Nie et al., 2012). However, to date, no small molecule-driven platform has demonstrated the ability to enhance reprogramming and support single-cell and FF culture of footprint-free iPSCs derived from human cells (Nichols and Smith, 2012). We show that the application of specific combinations of small molecule inhibitors in a stage-specific manner is key to enabling robust reprogramming and long-term culture. Studies have indicated that a reprogramming cell population is uniform in the early stages with major variations occurring thereafter (Polo et al., 2012). This suggests that counteracting lineage-specifying pathways early could facilitate the reprogramming process (Polo et al., 2012; Shu et al., 2013). Mesenchymal-to-epithelial transition (MET) has also been shown to support nuclear reprogramming (Li et al., 2010). By blocking differentiation cues early in the reprogramming process and promoting MET through small molecule inhibition of specific pathways, the efficiency of hiPSC generation was significantly improved using the FRM. Indeed, the use of the FRM resulted in robust reprogramming using episomal vectors, in FF and single-cell culture systems, including a demonstration of OCT4/SOX2/SV40LT-only episomal reprogramming.
Many ambitious hiPSC banking efforts are currently underway, with the goal of deriving populations of cells from different genetic and disease backgrounds (Rao, 2013). The derivation of footprint-free hiPSCs from various parental somatic sources in a multiplex manner has proven to be difficult and laborious (Narsinh et al., 2011; Saha and Jaenisch, 2009). By significantly improving the efficiency of episomal-induced reprogramming and selecting bona fide hiPSCs by direct flow cytometry sorting into 96-well plate formats at clonal density, cell line generation and characterization can be multiplexed. This system enables a single researcher to parallel reprogram multiple somatic lines using minimal starting cell material and derive many transgene-free hiPSC clones in single-cell, FF culture systems with minimal effort and in an expedited time frame.
Long-term passage of pluripotent stem cells in environments applicable to scaled cell production is hampered by spontaneous differentiation and genomic instability (Laurent et al., 2011; Taapken et al., 2011). Preventing spontaneous differentiation while increasing clonality with the use of small molecules has been demonstrated in both mouse and human pluripotent stem cells; however, the latter required ectopic expression of several pluripotency genes resulting in genetic manipulation (Hanna et al., 2010a; Ying et al., 2008). These cells represent the ground state of pluripotency that is associated with a capacity to give rise to all cell types and expression of unique genes associated with core pluripotency (Hanna et al., 2010b; Nichols and Smith, 2012). The more developmentally advanced stage of pluripotency representing the metastable state has been associated with human pluripotent stem cell cultures and is defined by properties that include spontaneous differentiation, high rate of cell death upon single-cell dissociation, and the inability to give rise to all somatic cell types (Nichols and Smith, 2012). We identified inhibition of TGF-β as a contributing factor in the spontaneous differentiation observed in long-term passage of transgene-free hiPSCs. FMM does not include a TGF-β inhibitor and is able to support the continuous maintenance of footprint-free hiPSCs passaged as single cells in a FF environment without the additional requirement of transgene expression. These media and culture systems facilitate culture practices for everyday users, such as removing the requirement for clump passaging, daily culture feeding, or culture cleaning, and therefore deliver a robust basis for a scaled manufacturing process, relevant for future cell therapy applications. We demonstrate that the genomic stability of multiple transgene-free hiPSC lines is supported by FMM after long-term single-cell enzymatic passages in a FF culture environment. This improved viability and stability in single-cell culture will be particularly useful in hiPSC applications such as selection of genomic modifications in disease-correction studies (Saha and Jaenisch, 2009). Collectively, our data suggest that hiPSCs cultured in FMM portray properties commonly associated with the ground state as demonstrated by features such as repression of differentiation gene expression and high clonality; however, additional studies will be necessary to confirm these observations (De Los Angeles et al., 2012; Hanna et al., 2010a, 2010b; Ying et al., 2008). The potential FMM-induced ground state is seen to be independent of transgene expression but also appears to be independent of derivation method, source of parental line, or previously maintained culture conditions. These qualities are lost when hiPSCs are switched from FMM to conventional culture (FMM→Conv) but gained when switched from conventional culture to FMM (Conv→FMM), suggesting that the ground state may be interconvertible as seen previously with mouse pluripotent stem cells (Marks et al., 2012). Two recent advancements in mouse iPSC technology include the elimination reprogramming factors in deriving mouse iPSCs and synchronizing the reprogramming process to nearly perfect efficiencies (Hou et al., 2013; Rais et al., 2013). The need for optimal culture conditions, i.e., 2i, was an important requirement for the success of both of these studies. FMM may represent the human pluripotent stem cell version of 2i and help accelerate the success of current studies. During the preparation of this manuscript, two studies demonstrated the maintenance of pluripotent stem cells in the ground state or resembling preimplantation epiblast using unique small molecule cocktails with similar pathway inhibition as utilized in FMM (Chan et al., 2013; Gafni et al., 2013).
The goal of the current study was to define a platform that enables hiPSC derivation and maintenance to the point where reprogramming is no longer a source of variability and/or gating activity for downstream cell line use. The FRM/FMM-multistage platform represents an advanced method for derivation and maintenance of hiPSCs that is amenable to all users. This study aims to serve as a resource and a blueprint for the derivation and maintenance of hiPSCs that are appropriate for industrial and clinical use.
Experimental Procedures
Animal Handling and Maintenance
Fate Therapeutics complies with all applicable provisions of the Animal Welfare Act and has established and maintained a program for activities involving animals in accordance with the NIH “Guide for the Care and Use of Laboratory Animals.”
hiPSC Maintenance in Small Molecule Culture
Derived hiPSCs were routinely passaged as single cells once confluency of the culture reached 75%–90%. Overconfluency was seen to exhaust the medium and result in differentiation. For single-cell dissociation, hiPSCs were washed once with PBS (Mediatech) and treated with Accutase (Millipore) for 3–5 min at 37°C followed with pipetting to ensure single-cell dissociation. The single-cell suspension was then mixed in equal volume with conventional medium, centrifuged at 225 × g for 4 min, resuspened in FMM, and plated on Matrigel-coated surface. Passages were typically 1:6–1:8, transferred tissue culture plates previously coated with Matrigel for 2–4 hr in 37°C or Vitronectin for 1 hr at 25°C and fed every 2–3 days with FMM. Cell cultures were maintained in a humidified incubator set at 37°C and 5% CO2. Medium formulations for FMM and FRM are described in Table S1. SMC4 culture is discussed previously (Valamehr et al., 2012).
Acknowledgments
We gratefully thank WiCell for karyotype analysis, copy number variation analysis, and insightful discussion and the University of California, San Diego, Histology Core Facility for tissue processing. All authors are current employees of Fate Therapeutics, Inc.
Footnotes
This is an open-access article distributed under the terms of the Creative Commons Attribution-NonCommercial-No Derivative Works License, which permits non-commercial use, distribution, and reproduction in any medium, provided the original author and source are credited.
Accession Numbers
The Gene Expression Omnibus (GEO) accession number for the Affymetrix profiling reported in this paper is GSE50868. The GEO accession number for the Array Comparative Genomic Hybridization and Single Nucleotide in this paper is GSE54080.
Supplemental Information
References
- Abujarour R., Valamehr B., Robinson M., Rezner B., Vranceanu F., Flynn P. Optimized surface markers for the prospective isolation of high-quality hiPSCs using flow cytometry selection. Sci. Rep. 2013;3:1179. doi: 10.1038/srep01179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beattie G.M., Lopez A.D., Bucay N., Hinton A., Firpo M.T., King C.C., Hayek A. Activin A maintains pluripotency of human embryonic stem cells in the absence of feeder layers. Stem Cells. 2005;23:489–495. doi: 10.1634/stemcells.2004-0279. [DOI] [PubMed] [Google Scholar]
- Chan Y.S., Göke J., Ng J.H., Lu X., Gonzales K.A., Tan C.P., Tng W.Q., Hong Z.Z., Lim Y.S., Ng H.H. Induction of a human pluripotent state with distinct regulatory circuitry that resembles preimplantation epiblast. Cell Stem Cell. 2013;13:663–675. doi: 10.1016/j.stem.2013.11.015. [DOI] [PubMed] [Google Scholar]
- De Los Angeles A., Loh Y.H., Tesar P.J., Daley G.Q. Accessing naïve human pluripotency. Curr. Opin. Genet. Dev. 2012;22:272–282. doi: 10.1016/j.gde.2012.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fusaki N., Ban H., Nishiyama A., Saeki K., Hasegawa M. Efficient induction of transgene-free human pluripotent stem cells using a vector based on Sendai virus, an RNA virus that does not integrate into the host genome. Proc. Jpn. Acad., Ser. B, Phys. Biol. Sci. 2009;85:348–362. doi: 10.2183/pjab.85.348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gafni O., Weinberger L., Mansour A.A., Manor Y.S., Chomsky E., Ben-Yosef D., Kalma Y., Viukov S., Maza I., Zviran A. Derivation of novel human ground state naive pluripotent stem cells. Nature. 2013;504:282–286. doi: 10.1038/nature12745. [DOI] [PubMed] [Google Scholar]
- Han D.W., Greber B., Wu G., Tapia N., Araúzo-Bravo M.J., Ko K., Bernemann C., Stehling M., Schöler H.R. Direct reprogramming of fibroblasts into epiblast stem cells. Nat. Cell Biol. 2011;13:66–71. doi: 10.1038/ncb2136. [DOI] [PubMed] [Google Scholar]
- Hanna J., Cheng A.W., Saha K., Kim J., Lengner C.J., Soldner F., Cassady J.P., Muffat J., Carey B.W., Jaenisch R. Human embryonic stem cells with biological and epigenetic characteristics similar to those of mouse ESCs. Proc. Natl. Acad. Sci. USA. 2010;107:9222–9227. doi: 10.1073/pnas.1004584107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanna J.H., Saha K., Jaenisch R. Pluripotency and cellular reprogramming: facts, hypotheses, unresolved issues. Cell. 2010;143:508–525. doi: 10.1016/j.cell.2010.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirata T., Amano T., Nakatake Y., Amano M., Piao Y., Hoang H.G., Ko M.S. Zscan4 transiently reactivates early embryonic genes during the generation of induced pluripotent stem cells. Sci. Rep. 2012;2:208. doi: 10.1038/srep00208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou P., Li Y., Zhang X., Liu C., Guan J., Li H., Zhao T., Ye J., Yang W., Liu K. Pluripotent stem cells induced from mouse somatic cells by small-molecule compounds. Science. 2013;341:651–654. doi: 10.1126/science.1239278. [DOI] [PubMed] [Google Scholar]
- Laurent L.C., Ulitsky I., Slavin I., Tran H., Schork A., Morey R., Lynch C., Harness J.V., Lee S., Barrero M.J. Dynamic changes in the copy number of pluripotency and cell proliferation genes in human ESCs and iPSCs during reprogramming and time in culture. Cell Stem Cell. 2011;8:106–118. doi: 10.1016/j.stem.2010.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee A.S., Tang C., Rao M.S., Weissman I.L., Wu J.C. Tumorigenicity as a clinical hurdle for pluripotent stem cell therapies. Nat. Med. 2013;19:998–1004. doi: 10.1038/nm.3267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R., Liang J., Ni S., Zhou T., Qing X., Li H., He W., Chen J., Li F., Zhuang Q. A mesenchymal-to-epithelial transition initiates and is required for the nuclear reprogramming of mouse fibroblasts. Cell Stem Cell. 2010;7:51–63. doi: 10.1016/j.stem.2010.04.014. [DOI] [PubMed] [Google Scholar]
- Lin T., Ambasudhan R., Yuan X., Li W., Hilcove S., Abujarour R., Lin X., Hahm H.S., Hao E., Hayek A., Ding S. A chemical platform for improved induction of human iPSCs. Nat. Methods. 2009;6:805–808. doi: 10.1038/nmeth.1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marks H., Kalkan T., Menafra R., Denissov S., Jones K., Hofemeister H., Nichols J., Kranz A., Stewart A.F., Smith A., Stunnenberg H.G. The transcriptional and epigenomic foundations of ground state pluripotency. Cell. 2012;149:590–604. doi: 10.1016/j.cell.2012.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa M., Koyanagi M., Tanabe K., Takahashi K., Ichisaka T., Aoi T., Okita K., Mochiduki Y., Takizawa N., Yamanaka S. Generation of induced pluripotent stem cells without Myc from mouse and human fibroblasts. Nat. Biotechnol. 2008;26:101–106. doi: 10.1038/nbt1374. [DOI] [PubMed] [Google Scholar]
- Narsinh K., Narsinh K.H., Wu J.C. Derivation of human induced pluripotent stem cells for cardiovascular disease modeling. Circ. Res. 2011;108:1146–1156. doi: 10.1161/CIRCRESAHA.111.240374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols J., Smith A. Pluripotency in the embryo and in culture. Cold Spring Harb. Perspect. Biol. 2012;4:a008128. doi: 10.1101/cshperspect.a008128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nie B., Wang H., Laurent T., Ding S. Cellular reprogramming: a small molecule perspective. Curr. Opin. Cell Biol. 2012;24:784–792. doi: 10.1016/j.ceb.2012.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Doherty R., Greiser U., Wang W. Nonviral methods for inducing pluripotency to cells. Biomed Res. Int. 2013;2013:705902. doi: 10.1155/2013/705902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okita K., Matsumura Y., Sato Y., Okada A., Morizane A., Okamoto S., Hong H., Nakagawa M., Tanabe K., Tezuka K. A more efficient method to generate integration-free human iPS cells. Nat. Methods. 2011;8:409–412. doi: 10.1038/nmeth.1591. [DOI] [PubMed] [Google Scholar]
- Pera M.F., Trounson A.O. Human embryonic stem cells: prospects for development. Development. 2004;131:5515–5525. doi: 10.1242/dev.01451. [DOI] [PubMed] [Google Scholar]
- Polo J.M., Anderssen E., Walsh R.M., Schwarz B.A., Nefzger C.M., Lim S.M., Borkent M., Apostolou E., Alaei S., Cloutier J. A molecular roadmap of reprogramming somatic cells into iPS cells. Cell. 2012;151:1617–1632. doi: 10.1016/j.cell.2012.11.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rais Y., Zviran A., Geula S., Gafni O., Chomsky E., Viukov S., Mansour A.A., Caspi I., Krupalnik V., Zerbib M. Deterministic direct reprogramming of somatic cells to pluripotency. Nature. 2013;502:65–70. doi: 10.1038/nature12587. [DOI] [PubMed] [Google Scholar]
- Rao M. iPSC crowdsourcing: a model for obtaining large panels of stem cell lines for screening. Cell Stem Cell. 2013;13:389–391. doi: 10.1016/j.stem.2013.09.005. [DOI] [PubMed] [Google Scholar]
- Saha K., Jaenisch R. Technical challenges in using human induced pluripotent stem cells to model disease. Cell Stem Cell. 2009;5:584–595. doi: 10.1016/j.stem.2009.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu J., Wu C., Wu Y., Li Z., Shao S., Zhao W., Tang X., Yang H., Shen L., Zuo X. Induction of pluripotency in mouse somatic cells with lineage specifiers. Cell. 2013;153:963–975. doi: 10.1016/j.cell.2013.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taapken S.M., Nisler B.S., Newton M.A., Sampsell-Barron T.L., Leonhard K.A., McIntire E.M., Montgomery K.D. Karotypic abnormalities in human induced pluripotent stem cells and embryonic stem cells. Nat. Biotechnol. 2011;29:313–314. doi: 10.1038/nbt.1835. [DOI] [PubMed] [Google Scholar]
- Takahashi K., Tanabe K., Ohnuki M., Narita M., Ichisaka T., Tomoda K., Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- Thomson J.A., Itskovitz-Eldor J., Shapiro S.S., Waknitz M.A., Swiergiel J.J., Marshall V.S., Jones J.M. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282:1145–1147. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- Valamehr B., Tsutsui H., Ho C.M., Wu H. Developing defined culture systems for human pluripotent stem cells. Regen. Med. 2011;6:623–634. doi: 10.2217/rme.11.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valamehr B., Abujarour R., Robinson M., Le T., Robbins D., Shoemaker D., Flynn P. A novel platform to enable the high-throughput derivation and characterization of feeder-free human iPSCs. Sci. Rep. 2012;2:213. doi: 10.1038/srep00213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., Yang J., Liu H., Lu D., Chen X., Zenonos Z., Campos L.S., Rad R., Guo G., Zhang S. Rapid and efficient reprogramming of somatic cells to induced pluripotent stem cells by retinoic acid receptor gamma and liver receptor homolog 1. Proc. Natl. Acad. Sci. USA. 2011;108:18283–18288. doi: 10.1073/pnas.1100893108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren L., Manos P.D., Ahfeldt T., Loh Y.H., Li H., Lau F., Ebina W., Mandal P.K., Smith Z.D., Meissner A. Highly efficient reprogramming to pluripotency and directed differentiation of human cells with synthetic modified mRNA. Cell Stem Cell. 2010;7:618–630. doi: 10.1016/j.stem.2010.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe K., Ueno M., Kamiya D., Nishiyama A., Matsumura M., Wataya T., Takahashi J.B., Nishikawa S., Nishikawa S., Muguruma K., Sasai Y. A ROCK inhibitor permits survival of dissociated human embryonic stem cells. Nat. Biotechnol. 2007;25:681–686. doi: 10.1038/nbt1310. [DOI] [PubMed] [Google Scholar]
- Xu Y., Zhu X., Hahm H.S., Wei W., Hao E., Hayek A., Ding S. Revealing a core signaling regulatory mechanism for pluripotent stem cell survival and self-renewal by small molecules. Proc. Natl. Acad. Sci. USA. 2010;107:8129–8134. doi: 10.1073/pnas.1002024107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying Q.L., Wray J., Nichols J., Batlle-Morera L., Doble B., Woodgett J., Cohen P., Smith A. The ground state of embryonic stem cell self-renewal. Nature. 2008;453:519–523. doi: 10.1038/nature06968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J., Vodyanik M.A., Smuga-Otto K., Antosiewicz-Bourget J., Frane J.L., Tian S., Nie J., Jonsdottir G.A., Ruotti V., Stewart R. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- Yu J., Hu K., Smuga-Otto K., Tian S., Stewart R., Slukvin I.I., Thomson J.A. Human induced pluripotent stem cells free of vector and transgene sequences. Science. 2009;324:797–801. doi: 10.1126/science.1172482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H., Wu S., Joo J.Y., Zhu S., Han D.W., Lin T., Trauger S., Bien G., Yao S., Zhu Y. Generation of induced pluripotent stem cells using recombinant proteins. Cell Stem Cell. 2009;4:381–384. doi: 10.1016/j.stem.2009.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H., Li W., Zhu S., Joo J.Y., Do J.T., Xiong W., Kim J.B., Zhang K., Schöler H.R., Ding S. Conversion of mouse epiblast stem cells to an earlier pluripotency state by small molecules. J. Biol. Chem. 2010;285:29676–29680. doi: 10.1074/jbc.C110.150599. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


