Figure 1.
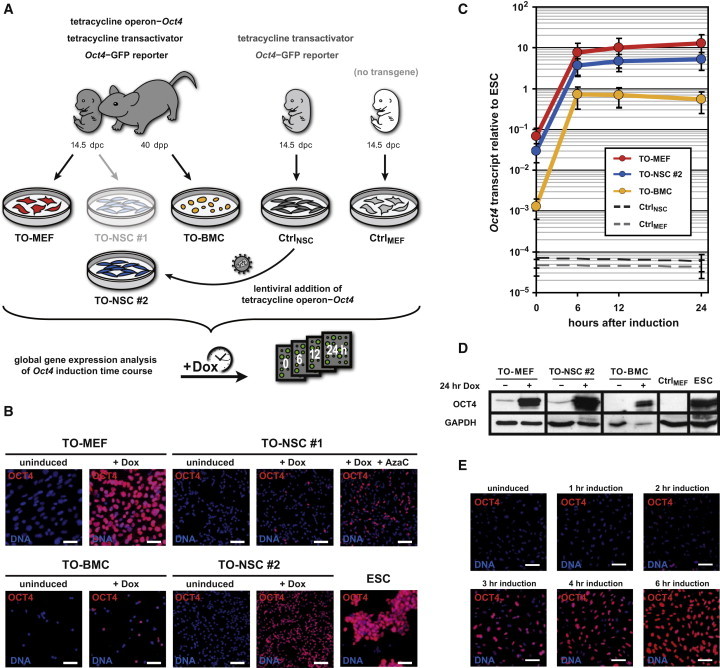
Generation and Characterization of Tetracycline-Inducible Oct4 Cells and Controls Used for Dynamic Global Gene-Expression Analysis
(A) Fibroblasts (TO-MEFs), neural stem cells (TO-NSCs number 1), and bone marrow cells (TO-BMCs) contain a tetracycline transactivator, a tetracycline-operon Oct4-expression cassette, and an Oct4-GFP reporter. NSCs with a tetracycline transactivator and an Oct4-GFP reporter but lacking a tetracycline-operon Oct4 cassette (CtrlNSC) served as a control for nonspecific transactivator activity. Wild-type C57BL/6 fibroblasts were used as a control for drug response artifacts (CtrlMEF). TO-NSCs number 2 were generated after transducing CtrlNSC with a lentiviral vector coding for a tetracycline-operon Oct4. Transcriptional effects of Oct4 expression were investigated by microarray analysis at different time points after doxycycline induction.
(B) Immunological staining for OCT4 protein (red) in the different cell types after 24 hr of doxycycline induction. Nuclei were counterstained with Hoechst (blue). The scale bars represent 50 μm.
(C) Expression of Oct4 mRNA (qRT-PCR), normalized to an embryonic stem cell (ESC) control. The error bars represent SE of calculations based on ΔCt values obtained from two different housekeeping genes: Gapdh and Actb (n = 1).
(D) OCT4 protein immunoblot of induced and uninduced samples. GAPDH was used as a loading control.
(E) OCT4 protein translation time course after doxycycline induction. OCT4 protein and nuclei are shown in red and blue, respectively. The scale bars represent 50 μm.
Abbreviations: dpc, days postcoitum; dpp, days postpartum; Dox, doxycycline; AzaC, 5-azacytidine; TO, tetracycline-inducible. See also Figure S1 as well as Tables S5 and S6.
