Figure 5.
Validation of OCT4 Target Genes Identified in the Microarray Analysis
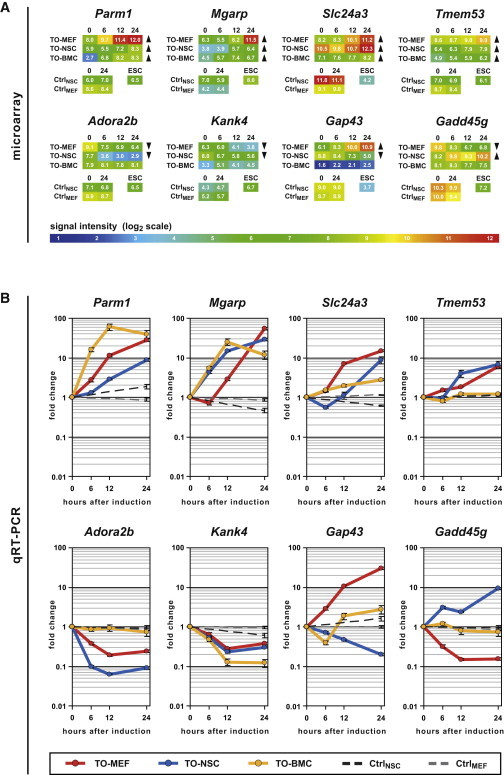
(A) The only four commonly upregulated (Parm1, Mgarp, Slc24a3, and Tmem53) and two examples of commonly downregulated (Adora2b and Kank4) as well as contrarily regulated genes (Gap43 and Gadd45 g) were chosen. Microarray signal intensities for all samples and controls at different time points after induction are shown as heatmaps. Arrows indicate in which cell types the criteria for up- or downregulation were fulfilled.
(B) Dynamic gene-expression analysis of the same genes using qRT-PCR, normalized to the respective uninduced cells. The error bars represent SE of calculations based on ΔCt values obtained from two different housekeeping genes: Gapdh and Actb (n = 1).
See also Table S5.
