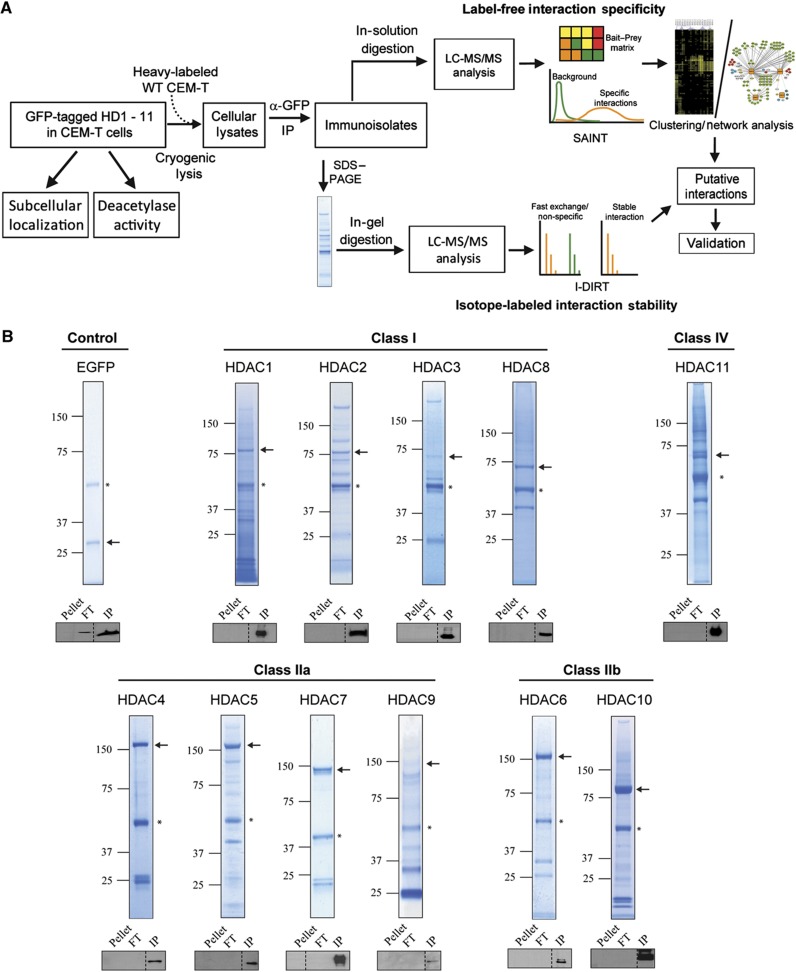
Figure 2.
Proteomic workflow and immunoisolation of 11 EGFP-tagged human HDACs. (A) Workflow for immunoisolation of HDACs from CEM T cell lines stably expressing HDAC–EGFP. HDAC–EGFP immunoisolates were subjected to label-free and isotope-labeled AP-MS workflows using SAINT and/or I-DIRT analysis, respectively. Hierarchical clustering and interaction networks visualized HDAC–HDAC and HDAC–prey relationships. Candidate protein interactions from global AP-MS were supported by molecular imaging and biochemical approaches. (B) Representative SDS–PAGE separations of Coomassie-stained EGFP-tagged HDAC1–11 immunoisolates. EGFP only immunoisolate is shown as a control. Arrows indicate the band containing the isolated bait. *, contaminant band. Western blotting assessed efficiency of HDAC–EGFP recovery in elution (IP) fraction relative to unbound flowthrough (FT) and insoluble cell pellet (pellet). Ten percent of each fraction was analyzed.