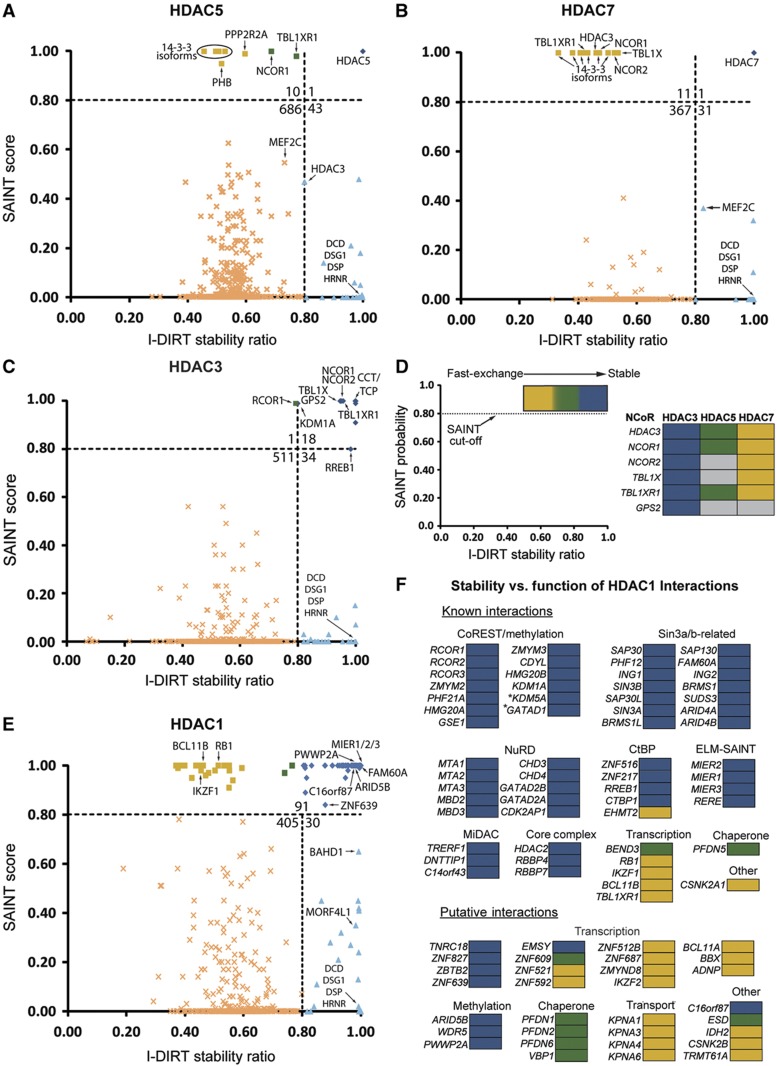
Figure 6.
Profiling of relative interaction stability within HDAC-containing complexes. Scatter plots show the relationship between interaction specificity (SAINT scores) and stability/specificity (I-DIRT ratios). Data shown are for common protein identifications between label-free and isotope-labeled AP-MS approaches from (A) HDAC5, (B) HDAC7, (C) HDAC3, and (E) HDAC1 isolations. Dashed lines represent selected thresholds and total protein number in each quadrant is shown. Selected data points are labeled with gene symbols. (D) Left, a region of high SAINT specificity, but varying I-DIRT ratios (0.5 to 1.0), is indicated by a color coded gradient indicating a stability range. Right, the relative stability of NCoR complex members is compared for HDAC3, 5, and 7. Gray boxes indicate the protein was absent or below SAINT threshold. (F) Known (top) and putative (bottom) HDAC1 interactions with SAINT scores>0.80 (n=90) are listed as gene symbols, depicted with their relative stability (D), and classified by known HDAC1 complexes or cellular function.