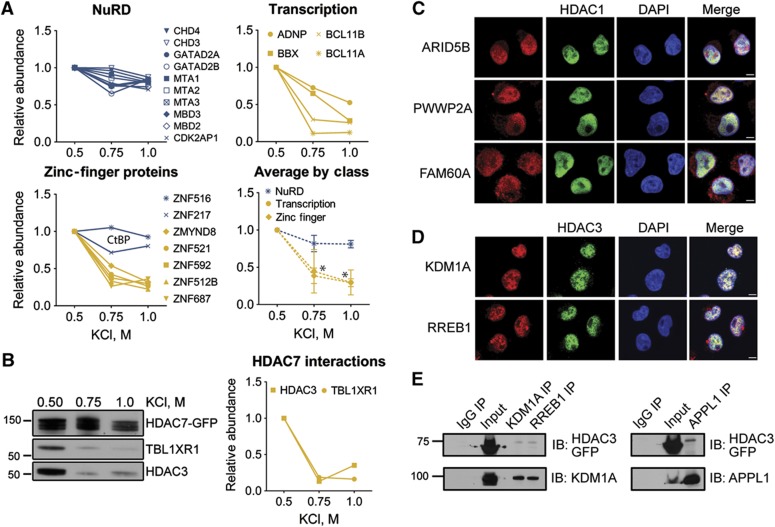
Figure 7.
Biochemical validation and confocal immunofluorescence confirm HDAC interactions identified by proteomics. (A) Biochemical validation of HDAC1 interaction stability. The relative abundance of HDAC1 interactions from immunoisolations performed under increasing salt concentration was determined for selected high (blue) and low (yellow) stability proteins comprising the NuRD complex (top left), transcription factors (top right), and zinc-finger proteins (bottom left). The average relative abundance (±s.d.) of these classes as a function of [KCl] (bottom right) is plotted, excluding ZNF518 and ZNF217 (CtBP complex members). Statistical significance of KCl-dependent relative abundances was assessed compared to the average NuRD-relative abundance (two-way ANOVA, *P<0.001). (B) Biochemical validation of HDAC7 interaction stability. The relative abundances of TBL1XR1 and HDAC3 were assessed by western blotting, normalized by densitometry to HDAC7. (C) Colocalization of HDAC1–EGFP with ARID5B, PWWP2A, and FAM60A, and (D) colocalization HDAC3–EGFP with KDM1A and RREB1 in CEM T-cell lines. Localization of EGFP-tagged HDACs and selected proteins were detected using anti-GFP antibody (green) and antibodies against endogenous proteins (red); DNA is visualized by DAPI (blue); × 63 oil immersion lens; scale bar 5 μm. (E) Reciprocal affinity purifications (IP) for HDAC3–EGFP using antibodies against endogenous KDM1A and RREB1 (left), and APPL1 (right). EGFP-tagged HDAC3 was detected by western blot. IgG was used as negative control.