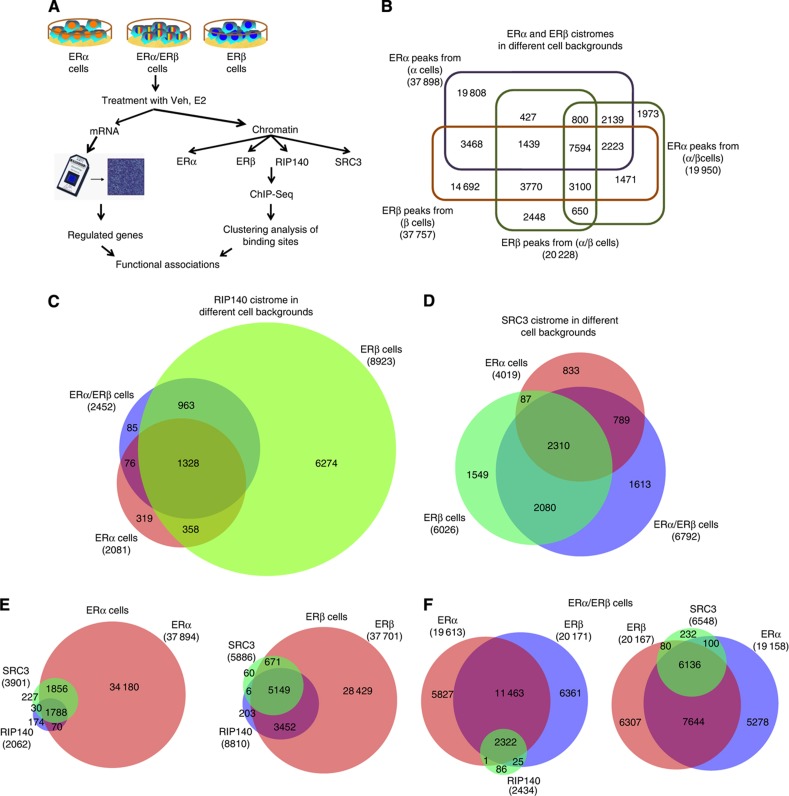
Figure 1.
Genome-wide analysis of ERα, ERβ, SRC3 and RIP140 chromatin binding by ChIP-seq. (A) Experimental design for ChIP-seq, gene microarray, and clustering analyses. Veh, vehicle; E2, 10 nM 17β-estradiol. (B) Comparison of ERα- and ERβ-binding sites in the different cell backgrounds. (C) Comparison of RIP140-binding sites in the different cell backgrounds. (D) Comparison of SRC3-binding sites in the different cell backgrounds. (E) Comparison of the number of ER and coregulator (SRC3, RIP140)-binding sites in ERα cells or ERβ cells. (F) Overlap of RIP140- or SRC3-binding sites with ERs in ERα/ERβ cells.