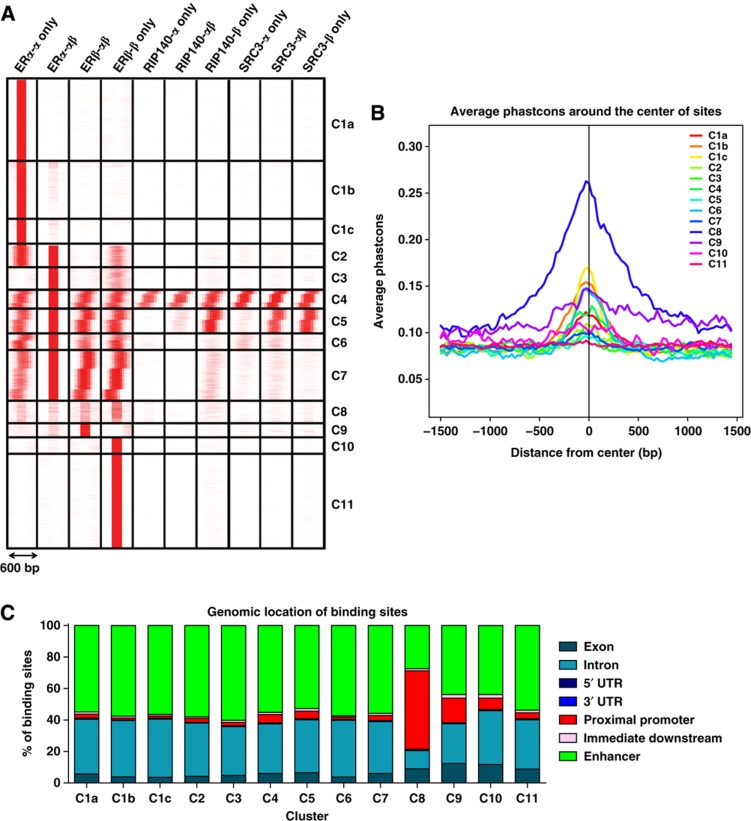
Figure 2.
Categorization of binding sites into clusters based on ERα, ERβ, SRC3, and RIP140 cistromes. (A) Clustering of the binding sites. seqMINER software was used for clustering based on colocalization of different factors in different cell backgrounds within a 300-bp window in both directions. (B) Conservation of binding sites among vertebrates: cistrome conservation tool was used to compare conservation of binding sites from different clusters among vertebrates. (C) Genomic location of binding sites: web-based CEAS tool was used for identifying genomic location of binding sites from each cluster.