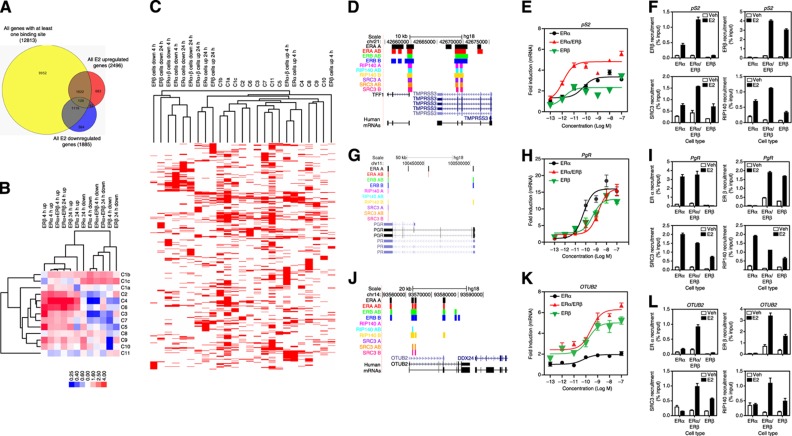
Figure 4.
Association of ER-mediated gene expression with receptor and coregulator cistromes. (A) Comparison of number of genes with at least one binding site within 20 kb and E2-upregulated or -downregulated genes. (B) Association of gene regulations in different cell backgrounds with binding site clusters: enrichment of binding-site clusters (C1–C11) associated with upregulated and downregulated genes in each cell background after 4 or 24 h of estradiol treatment is shown. Level of enrichment is designated by the color scale. Cluster 3.0 was used to cluster the data. Data were visualized using Java Treeview. (C) Clustering of genes that are regulated by E2 treatment and genes that have binding sites from different clusters: BED files for E2-regulated genes were generated using Genespring. The seqMINER enrichment based method was used to calculate the density array for the E2-regulated gene BED files and BED files obtained for each cluster from Figure 2A. Density array was clustered using Cluster 3.0 and data was visualized using Java Treeview. (D) The pS2 gene and associated binding sites for ERα, ERβ, RIP140, and SRC3 in the three cell backgrounds. (E) The pS2 gene regulation by E2 (10−13 to 10−7 M) in the different cell backgrounds. (F) The pS2 gene ChIP for ERα, ERβ, and coregulators in cells treated with control vehicle (0.01% ethanol) or 10 nM E2. (G) The PgR gene and associated binding sites in cells treated with control vehicle (0.01% ethanol) or 10 nM E2. (H) The PgR gene regulation by E2 in the different cell backgrounds. (I) The PgR gene ChIP for ERα, ERβ, and coregulators in cells treated with control vehicle (0.01% ethanol) or 10 nM E2. (J) The OTUB2 gene and associated binding sites in cells treated with control vehicle (0.01% ethanol) or 10 nM E2. (K) The OTUB2 gene regulation by E2 in the different cell backgrounds. (L) The OTUB2 gene ChIP for ERα, ERβ, and coregulators in cells treated with control vehicle (0.01% ethanol) or 10 nM E2.