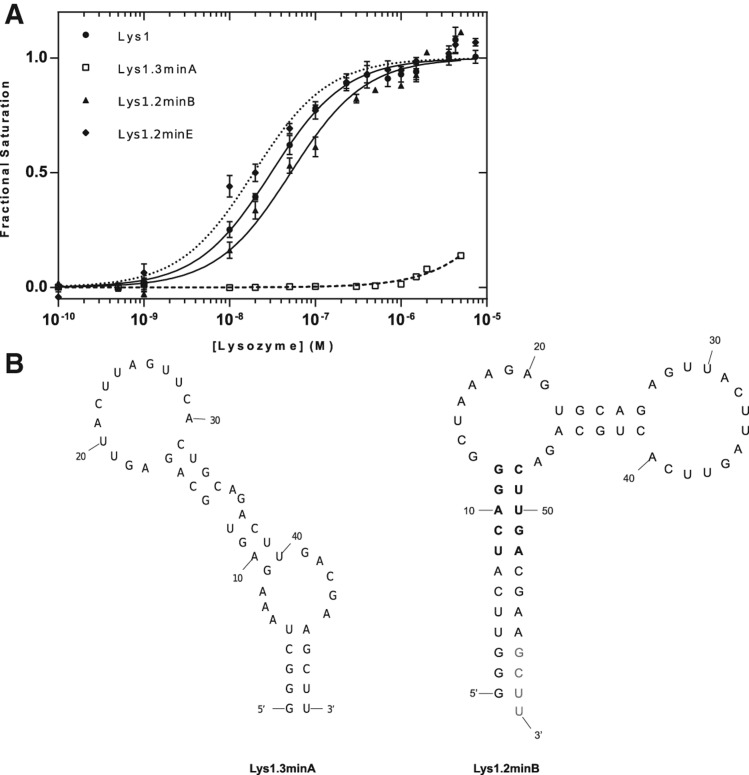
FIGURE 2.
Binding affinities for Lys1 constructs. (A) Binding affinities of full-length aptamer and deletion variants based on footprinting data. (B) Minimization of Lys1 using the predicted secondary structure 1.3 (Lys1.3minA) and secondary structure 1.2 (Lys1.2minB). Residues in gray were changed to CCC to form a thermodynamically stable stem in the Lys1.2minE construct. Residues in bold compose the five Watson-Crick based stems of Lys1.2minF, where UCAGG was replaced with GGGCG and AGUUC was replaced with CCCGC.