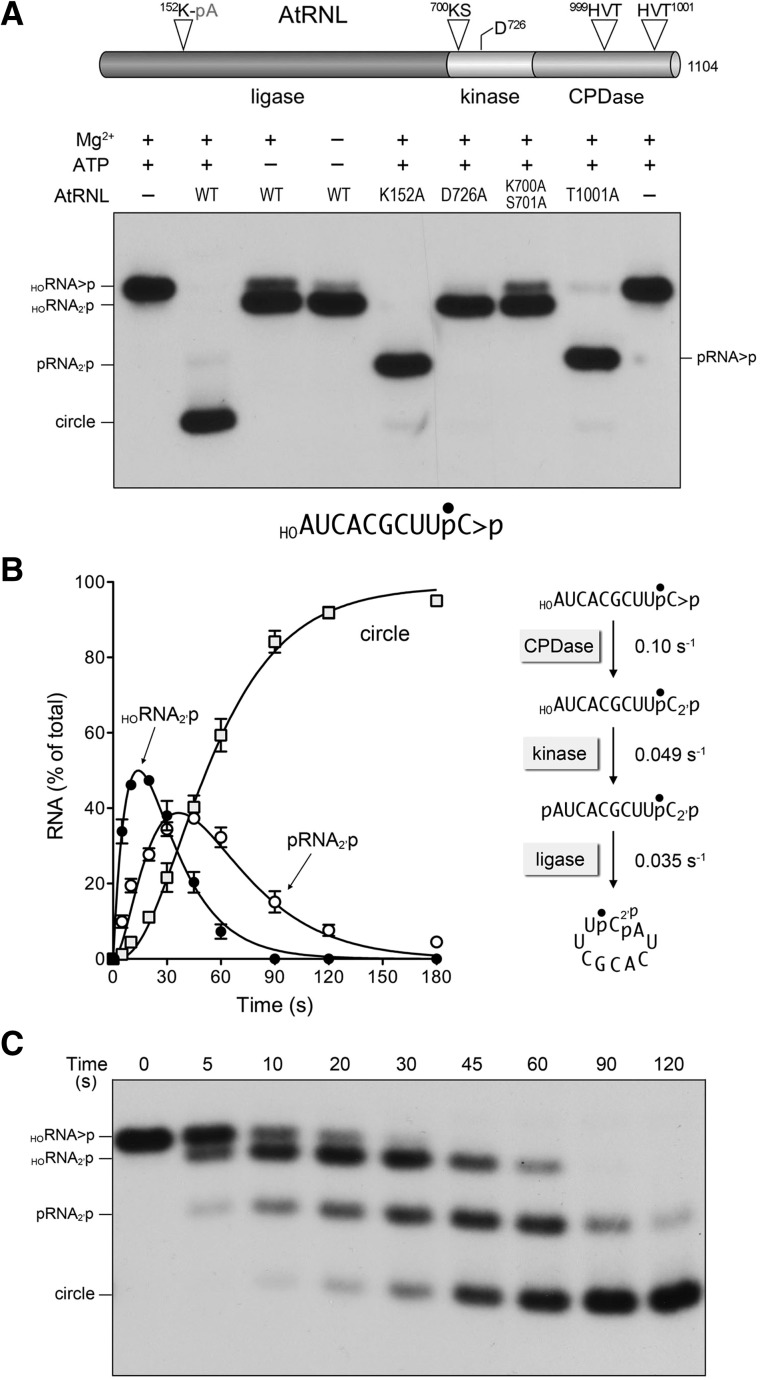
FIGURE 1.
One-pot assay for monitoring the AtRNL end-healing and sealing reactions. (A) Arabidopsis tRNA ligase is composed of an N-terminal ligase module, a central 5′ kinase module, and a C-terminal CPDase module, depicted as a cartoon model at the top of the panel. The site of covalent adenylylation at the ligase active site, the P-loop motif and aspartate general base at the kinase active site, and the two HxT motifs that comprise the CPDase active site are highlighted. AtRNL mutants with inactivating alanine substitutions in each of the three catalytic modules were reacted with a 3′ 32P-labeled 10-mer HORNA>p substrate (depicted at the bottom of the panel with the 32P-label denoted by •). Reaction mixtures (20 µL) containing 50 mM Tris-HCl, pH 8.0, 2 mM DTT, 10 mM MgCl2, 100 µM ATP, 20 nM 10-mer HORNA>p, and 1 µM AtRNL (wild type or mutant as specified) were incubated for 5 min at 22°C. Individual reaction components were included (+) or omitted (−) as specified. The reactions were quenched with an equal volume of 90% formamide, 30 mM EDTA. The products were analyzed by electrophoresis through a 20% polyacrylamide gel containing 7 M urea in TBE. An autoradiograph of the gel is shown. The position and identities of the radiolabeled RNA substrate and the various healed or sealed products are indicated on the left and right. (B,C) Order and kinetic profile of the splicing reactions under single-turnover conditions. A reaction mixture (200 µL) containing 50 mM Tris-HCl, pH 8.0, 50 mM NaCl, 2 mM DTT, 10 mM MgCl2, 0.1 mM ATP, 20 nM 10-mer HORNA>p substrate, and 1 µM AtRNL was incubated at 22°C. Aliquots (20 µL) were removed at the times specified and quenched immediately with an equal volume of 90% formamide, 30 mM EDTA. The time 0 sample was withdrawn and quenched prior to adding AtRNL. The products were resolved by urea-PAGE. An autoradiogram of the gel is shown in panel C. The levels of the healed HORNA2′p and pRNA2′p intermediates and the ligated circle product were quantified by scanning the gel with a Fujix BAS2500 imager and are plotted as a function of time in panel B. Each datum is the average of three separate time-course experiments ±SEM. The data were fit by nonlinear regression in Prism to a sequential three-step reaction pathway shown at right in panel B. The apparent rate constants for the CPD, kinase, and ligase reactions are indicated.