It is well known that metal ions play essential roles in RNA folding and catalysis. This study examines in detail metal ions bound to specific regions of a catalytic group II intron. These analyses based on crystallographic data provide new insight into how metal ions are localized in complex folded RNA structures.
Keywords: splicing, X-ray crystallography, tetraloop-receptor, T-loop, GA imino mismatch, ribonucleoprotein
Abstract
Metal ions promote both RNA folding and catalysis, thus being essential in stabilizing the structure and determining the function of large RNA molecules, including group II introns. The latter are self-splicing metalloribozymes, containing a heteronuclear four-metal-ion center within the active site. In addition to these catalytic ions, group II introns bind many other structural ions, including delocalized ions that bind the RNA diffusively and well-ordered ions that bind the RNA tightly with high occupancy. The latter ions, which can be studied by biophysical methods, have not yet been analyzed systematically. Here, we compare crystal structures of the group IIC intron from Oceanobacillus iheyensis and classify numerous site-bound ions, which are primarily localized in the intron core and near long-range tertiary contacts. Certain ion-binding sites resemble motifs observed in known RNA structures, while others are idiosyncratic to the group II intron. Particularly interesting are (1) ions proximal to the active site, which may participate in splicing together with the catalytic four-metal-ion center, (2) organic ions that bind regions predicted to interact with intron-encoded proteins, and (3) unusual monovalent ions bound to GU wobble pairs, GA mismatches, the S-turn, the tetraloop-receptor, and the T-loop. Our analysis extends the general principles by which ions participate in RNA structural organization and it will aid in the determination and interpretation of future RNA structures.
INTRODUCTION
Ions play a central role in all aspects of RNA biochemistry (Pyle 2002; Draper 2004, 2013; Leipply et al. 2009). Magnesium and potassium are the predominant metal ions bound to RNA molecules in vivo, because of their natural abundance and chemical properties (Feig and Uhlenbeck 1999). However, RNA can bind many more types of ions, including organic ions and alkaline, alkali-earth, and transition metals (Feig and Uhlenbeck 1999; DeRose 2003; Auffinger et al. 2011). RNA-bound ions can be studied using many informative biochemical and biophysical techniques. X-ray crystallography is useful because it provides simultaneous high-resolution information on specific ions and on the overall structure of the surrounding RNA molecule. To date, 36 different types of metals have been observed to bind RNA by X-ray crystallography (Auffinger et al. 2011; Schnabl et al. 2012).
Group II introns are among the largest RNA molecules of known structure (Toor et al. 2008a; Pyle 2010; Marcia et al. 2013b) and they are highly dependent on metal ions for their function (Sigel et al. 2000; Gordon and Piccirilli 2001; Kruschel and Sigel 2008). Magnesium is required for group II intron folding (Swisher et al. 2002; Su et al. 2003, 2005; Erat and Sigel 2007) and splicing (Podar et al. 1995; Gordon and Piccirilli 2001), while monovalent ions also facilitate these processes (Basu et al. 1998; Conn et al. 2002; Pyle et al. 2007; Lambert et al. 2009). In addition, monovalent ions dictate the splicing reaction pathway (branching vs. hydrolysis) (Jarrell et al. 1988; Daniels et al. 1996), and they play a key role in modulating alternative conformations of the ribozyme (Marcia and Pyle 2012). However, a systematic structural characterization of intron-bound ions has been missing due to the limited resolution of available crystal structures. In addition, the small number of structures solved in the presence of anomalous scattering ions did not allow for an unambiguous identification of all metal sites (Toor et al. 2008a, 2010). However, we recently succeeded in obtaining high-resolution structures of the Oceanobacillus iheyensis group II intron (OiGIIi) in numerous different ionic combinations that include diverse anomalous scattering ions (Li+/Mg2+, Na+/Mg2+, K+/Mg2+, Rb+/Mg2+, Cs+/Mg2+, Tl+/Mg2+, NH4+/Mg2+, K+/Ca2+, and K+/Ba2+) (Marcia and Pyle 2012; Marcia et al. 2013b). Using a combination of crystallographic techniques, we have identified and characterized numerous intron-bound ions, thereby providing us with a unique opportunity to examine the specificity of ion-binding sites in a complex RNA tertiary structure.
Descriptions of RNA-bound ions have been reported previously for other large RNAs. For instance, a study using Rb+, K+, Na+, and NH4+ led to the analysis of 204 ions in the 23S rRNA (Klein et al. 2004). A study using Pb2+, Sm3+, Gd3+, Yb3+, and Os(III) hexamine characterized 13 ions in RNase P (Kazantsev et al. 2009), and a study using Yb3+, Tb3+, Eu3+, Mn2+, and Tl+ analyzed 18 ions in the group I intron (Stahley et al. 2007). By comparison with previous reports, the present study on a group II intron is uniquely comprehensive. We have utilized a larger series of ions than previous studies, so the work provides new insights into the binding specificity of diverse ions within specific RNA motifs. Additionally, we solved each of our structures in the presence of a single monovalent and a single divalent ion type, while previous work involved complex combinations of ions, which inevitably translates into a more complex interpretation of the ion-binding sites and of their occupancies. Perhaps most importantly, unlike previous studies, we selected ions that are good mimics of the physiological ions potassium and magnesium, as demonstrated by the fact that many of them support intron splicing (Marcia and Pyle 2012).
Our study is obviously limited to the characterization of “site-bound” (or “chelated”) (Draper 2004) ions, which have been calculated to represent only a fraction of the total ions bound to an RNA molecule (Draper 2004; Freisinger and Sigel 2007). It can be expected that many more ions surround each intron molecule. The majority of these ions, which are crystallographically invisible, probably associate with the RNA diffusively and contribute to charge neutralization in a nonspecific manner (Abramovitz et al. 1996; Feig and Uhlenbeck 1999; Banatao et al. 2003; Draper 2004). In contrast, most of the ions observed crystallographically appear to bind the RNA site-specifically, reflecting high order and occupancy. Among these ions, those bound to the catalytic site and involved in splicing include a heteronuclear four-metal-ion center (Marcia and Pyle 2012; Marcia et al. 2013b) and other ions that we describe here for the first time. In addition, the ions bound to peripheral regions include all of the major RNA metal-binding sites that have been classified previously, as annotated in the database MeRNA (Stefan et al. 2006), i.e., GU wobble pairs (Cate and Doudna 1996; Klein et al. 2004), GA mismatches (Pley et al. 1994), magnesium clamps (Ennifar et al. 1999), AA-platforms (Basu et al. 1998), and G-phosphates (Klein et al. 2004). Interestingly, within the group II intron structures, some of these motifs display different binding selectivity and different geometry from that reported in other RNA structures. Finally, as observed in other RNAs, organic ions, such as anionic sulfonate groups (Kieft et al. 2010) and polyamines like spermine (Quigley et al. 1978), bind to the intron, too. Within the group II intron, it is significant that these molecules are clustered at sites predicted to bind partner proteins, such as the intron-encoded maturase. Taken together, this classification of intron-bound ions brings new sophistication to our knowledge of metal ion-binding sites in complex RNAs and helps establish first principles for understanding the role of metals in RNA structures.
RESULTS AND DISCUSSION
Ion assignments
All crystals of OiGIIi used in this work grew in space group P212121, with similar unit cell dimensions (a = 89.3 Å ± 0.6%, b = 95.6 Å ± 0.5%, and c = 225.4 Å ± 0.5%) and resolution of diffraction (∼3 Å, with the exception of crystals grown in Ba2+, which diffracted to 4 Å), independent of the construct used (Oi5eD1-5 or OiD1-5) (Marcia and Pyle 2012). Also, the corresponding structures are very similar to each other, with overall reciprocal RMSD values of <2 Å (Marcia and Pyle 2012). The structural similarity is indicative of the good adaptability of OiGIIi to the different ionic combinations used in this study (Marcia et al. 2013a) and guarantees that the structural comparison is not biased by differences in crystal packing or in RNA-backbone conformations. Despite the similarity of all structures, interpreting ion-binding sites at 3 Å resolution is difficult, especially considering the analogous scattering properties of some solvent molecules (Holbrook et al. 1978). In our work, we assigned ions to positive peaks of non-nucleotide electron density in 2Fo-Fc and Fo-Fc Fourier difference maps according to the following principles.
First, anomalous scattering Fourier difference maps were calculated as described (Marcia and Pyle 2012) for structures that contained heavy metals and these were used to assign the corresponding metal binding sites (Rb+ in PDB id. 4E8P; Tl+ in PDB id. 4E8Q; Cs+ in PDB id. 4E8R; and Ba2+ in PDB id. 4E8V). Based on the position of Tl+ and Rb+, which possess similar ionic properties to K+ (Auffinger et al. 2011), and considering that K+ typically displays distances of about 2.8–3.5 Å from coordinating ligands (Harding 2001, 2002; Mahler and Persson 2012), potassium-binding sites were assigned in the remaining potassium-containing structures (PDB id. 4E8K, 4E8M, 4E8T, 4FAQ, 4FAR, 4FAW, and 4FB0). Ammonium-binding sites were assigned using a similar rationale in the ammonium structure (PDB id. 4E8N), considering the similarity of NH4+ to K+ (Yamada et al. 1998; Auffinger et al. 2011).
After assigning these monovalent ion positions, the remaining non-nucleotide electron density peaks in the K+, Rb+, Tl+, Cs+, and NH4+ structures were assigned to magnesium if the binding sites displayed octahedral coordination geometry and were located at 2.0–2.4 Å from at least one coordinating ligand (Harding 2001, 2002; Erat and Sigel 2008; Auffinger et al. 2011). If these conditions were not met, electron density peaks were assigned to water molecules when they were of appropriate intensity and at hydrogen-bonding distance from suitable coordinating atoms. In all other cases the sites were left unoccupied. Similar considerations were used to assign the Ca2+-binding sites in the calcium structures (PDB id. 4E8K, 4E8T, and 4FAQ). Similarly, the positions of divalent ions and water molecules were used to assign magnesium and water sites in the Na+ and Li+ structures (4FAX and 4FAU, respectively). The refinement of these latter structures was then completed by the addition of monovalent ions where suitable density peaks remained unoccupied.
Finally, spermine, which was an essential component of the crystallization buffer, was tentatively modeled at two structural sites that displayed elongated electron density peaks. While the identity of these molecules cannot be conclusively assigned, the presence of polyamines at these positions is not only compatible with the electron density signal, but also with a characteristic set of hydrogen bonds and ionic interactions with neighboring RNA segments and with data from other nucleic acid structures (Quigley et al. 1978; Korolev et al. 2002). Similarly, 2-[4-(2-hydroxyethyl)piperazin-1-yl]ethanesulfonic acid (HEPES), which was used to buffer the crystallization solutions, was tentatively modeled at six sites. At these sites, weak anomalous scattering Fourier electron density peaks were present in the Cs+ and/or Ba2+ structures. Such peaks are compatible with the presence of the sulfur atom of HEPES because these diffraction data sets were collected at low X-ray energies (8343 and 8349 eV, respectively) at which sulfur displays appreciable anomalous scattering (f ″ = 0.52 electrons). The distribution of the electrostatic potential at these sites is also compatible with the binding of a zwitterionic molecule like HEPES, because one can identify regions of more negative potential that can bind the 2-hydroxyethyl-piperazin tail, which carries a positive charge on the N1 atom, and regions of less negative potential that can bind the anionic sulfonate head group, positioning the sulfur atom at ∼3.8 Å from neighboring nitrogen or oxygen atoms in the RNA. This binding mode resembles the binding of sulfate ions (Auffinger et al. 2004) or 2-(N-morpholino)-ethanesulfonic acid (MES) (Klein and Ferre-D'Amare 2006) observed in other RNA structures.
After modeling all ions, their occupancy was adjusted based on evaluation of solvent B-factors and on consistency with the B-factors of surrounding nucleotides.
Using the above strategy, we identified a total of 74 putative ion-binding sites (Fig. 1; Supplemental Table S1). Of these sites, 34 represent divalent metal ion-binding sites (M sites), 32 represent monovalent metal ion-binding sites (K sites), two are putative polycations (S sites), and six are putative zwitterions (A sites). Each site is named with an alphanumeric code, in which the letter identifies the ion type (M, K, S, or A) and precedes an Arabic numeral. We note that among these 74 sites, 11 display heavy metal binding but do not show corresponding cognate electron density at the same positions in structures obtained in Mg2+ and K+ (native conditions). Therefore, these ions were not modeled in the structures containing native ions. Presumably there is differential affinity between heavy and native metals at certain types of sites (Stahley et al. 2007). For this reason, these ions are not discussed extensively in the text, although they are reported in Figure 1 and Supplemental Table S1.
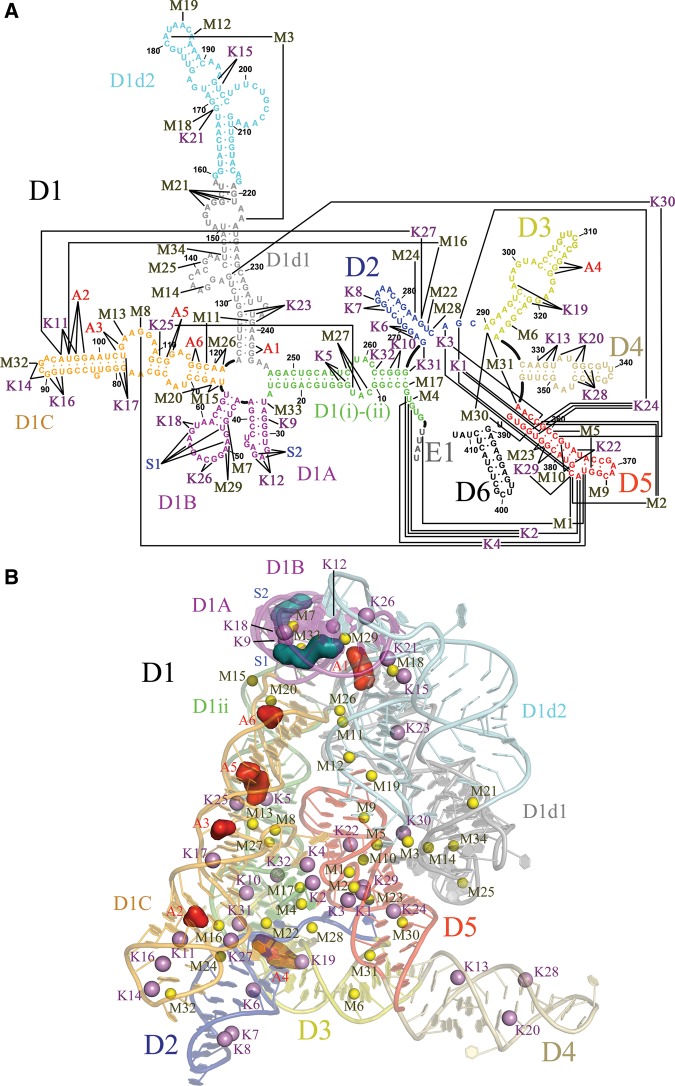
FIGURE 1.
Site-bound ions in O. iheyensis group II intron. (A) Secondary structure map of the crystallization construct adapted from Pyle (2010) to indicate the site-bound ions described in this work. D6 is included for completeness, but it was not present in the crystallization construct and its structure is currently undetermined. E1 indicates the four nucleotides of the 5′-exon. (B) Overall crystal structure in which all site-bound ions are represented as spheres. M sites are yellow, K sites violet, S sites blue, and A sites red. Each intron subdomain is depicted in a different color.
Overall ionic distribution
There are trends in the distribution of ion types throughout the intron structures. Ions bind to every intron domain and subdomain, resulting in 120 out of 390 intron residues in direct contact with at least one site-bound ion (Fig. 1A). However, we observe a different distribution of metallic vs. organic ions (Fig. 1B). Metallic ions—including both mono and divalent ions—dominate within the intron core. They are particularly concentrated within the active site, which has a high density of negative charges. In this region, three groups of ions can be identified. Within the active site, the heteronuclear four-metal ion center M1-M2-K1-K2 is in direct contact with the reactants and promotes splicing (Marcia and Pyle 2012). Around this center, other ions (M3, M5, K4, and possibly K3) come in direct contact with active-site residues, but not with the reactants themselves, thereby influencing splicing indirectly (Boudvillain and Pyle 1998). At a further distance from the active site, a wider network of ions (M4, M9, M10, M17, M23, K30, and possibly K22, K24, and K29) bind residues flanking active site elements, but their involvement in splicing has not been demonstrated to date. Additionally, mono- and divalent metal ions are localized near important long-range tertiary interactions outside the intron core. Interestingly, divalent ions are localized predominantly in D1 (M7-8, M11-15, M18-21, M25-27, M29, M32-34). This observation correlates with the common observation that divalent ions stabilize formation of tertiary interactions in nucleic acids and with the fact that D1 is the nucleation point for group II intron folding (Su et al. 2005; Pyle et al. 2007; Waldsich and Pyle 2008; Donghi et al. 2013). Monovalent ions are abundant in peripheral elements, where they interact with helical stems of D1C, D2, and D4 (K5–8, K11, K13–14, K16–17, K20, K25, K27–28). Fewer monovalent ions are also directly involved in long-range tertiary interactions (K9, K12, K15, K19, K23, and K26). Finally, organic ions (A and S sites) predominantly bind peripheral regions that are directly accessible from the bulk solvent and that are clustered on the face of the intron formed by D1A, D1B, and D1C (Fig. 1). Considering that most of these regions were shown to crosslink with proteins that are bound by group II introns (Matsuura et al. 2001; Dai et al. 2008), the organic ion-binding sites identified here may correspond to RNA interfaces that evolved to bind proteins. Specific structural features of all the various sites are discussed in the following sections.
Metal ions within the catalytic core
The catalytic core of the group II intron is formed by conserved motifs that include the catalytic triad (in D5), the 2-nt bulge (in D5), the J2/3 junction (connecting D2 and D3), and the 5′-splice junction connecting the 5′-end of the molecule (in D1) to the 5′-exon (Fig. 2; Boulanger et al. 1995; Schmidt et al. 1996; Costa et al. 2000; de Lencastre et al. 2005; de Lencastre and Pyle 2008; Marcia and Pyle 2012). As described above, metal ions in and around the catalytic core can be classified into three groups based on their distance from the splicing reactants.
FIGURE 2.
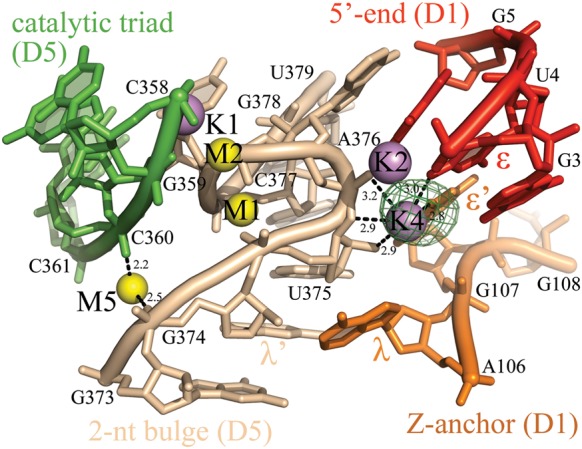
K4 and M5 bind the 2-nt bulge and the Z-anchor. M-sites are depicted as yellow spheres, K-sites as violet spheres, and the intron residues as sticks, color-coded by structural motif. The λ–λ′ and the ɛ–ɛ′ interactions are indicated. Anomalous difference Fourier electron density map at the K4 site is shown as a green mesh at 4.0 σ for Tl+ (PDB id. 4E8Q). At the M5 site, an anomalous difference Fourier electron density peak is visible for Ba2+ (PDB id. 4E8V, data not shown), but not for any monovalent ions. The M1-M2-K1-K2 catalytic center is indicated for reference. Inner-sphere coordination bonds are shown for K4 and M5 as black dashed lines and the corresponding distances are indicated in angstroms.
In direct contact with the reactants, M1, M2, K1, and K2 form the catalytic heteronuclear metal center that was described previously (Marcia and Pyle 2012). In this center, M1 and M2 form the binding platform for the RP nonbridging phosphoryl oxygen of the scissile phosphate and they play a key role in the chemistry of splicing. K1 modulates the toggling between active and inactive intron conformations throughout the splicing cycle. K2 bridges the 5′-end of the intron and the 2-nt bulge (A376 in D5) and it interacts with the hydrolyzed 5′-phosphate of the intron after the first step of splicing (Marcia and Pyle 2012).
Surrounding this core, an important set of additional ions is in contact with active site residues, but not with reactants. For example, M3 is located near the coordination and κ loops, and is clamped between the RP phosphoryl oxygen of A181 (EBS1, D1d2) and the SP phosphoryl oxygen of A223 (EBS3, D1d1), as described previously (Toor et al. 2008b; Supplemental Fig. S1). In this way, M3 may provide favorable stabilization for the two primary exon-binding sites (EBS1 and EBS3) perhaps enabling them to stack and orient themselves for reaction within the active site. K4 is a very dehydrated site that forms a dinuclear center with K2, sharing U4 and A376 as common ligands. K4 connects the 5′-end of the intron (O4 of U4, ɛ) with nucleotides in the Z-anchor (O6 of G107, ɛ′, D1C) and in the 2-nt bulge of D5 (O2′ and O3′ of U375, and the RP nonbridging phosphoryl oxygen of A376). M5 forms a magnesium clamp between the SP phosphoryl oxygen of C360 (catalytic triad) and the RP phosphoryl oxygen of G374 (λ). Together, K4 and M5 may contribute to the stabilization of the characteristic elbow shape adopted by D5 around the 2-nt bulge, which is essential for active site formation (Fig. 2). Finally, K3 is also localized within the catalytic core, but its identity and functional role are less evident (Marcia and Pyle 2012). K3 displays anomalous signal for Rb+, Tl+, and Cs+ and is in contact with the RP phosphoryl oxygen of C358 (catalytic triad), with O5′ and the SP phosphoryl oxygen of C377 (2-nt bulge), and with O3′ of A376 (2-nt bulge). K3 is also close to A287 (J2/3 junction, γ), C289 (J2/3 junction), and K1. Considering that the γ–γ′ interaction forms during the second step of splicing, K3 may play a particularly important role near the end of the splicing cycle (see Supplemental Fig. S2 in Marcia and Pyle 2012).
Moving outward from the catalytic center, a number of ions appear to interact with nucleobases that flank active site residues (M4, M9–10, M17, M22–23, M28, K22, K24, K29, and K30) (Supplemental Table S1).
A common feature of ions within the catalytic core (M1, M2, M3, M5, K1, K2, and K4) is that they are all present, with good occupancy, in both the pre- and the post-hydrolytic splicing steps (structures 4FAQ/4E8K and 4FAR/4FAW, respectively). This is consistent with previous phosphorothioate and NAIM interference experiments on the group II ai5γ intron, which revealed a network of functional ligands for these same ions, such as the RP phosphoryl oxygen atoms of G374 and of A376 (Boudvillain and Pyle 1998). These structural and functional observations strongly suggest that, in addition to the M1-M2-K1-K2 metal center, the concerted actions of M3, M5, K4, and possibly K3, play a decisive role in determining group II intron reactivity. In contrast, in structures that represent the toggled intermediate state, which is hypothesized to occur between the two steps of splicing (structures 4FAX and 4FAU) (Marcia and Pyle 2012), M1, M2, M3, M5, K1, K2, and K4 are not occupied.
Cations bound to helical motifs
Metal ions are often bound to helical stems within RNA structures, in which they either bridge phosphate groups of consecutive nucleotides or interact with nucleobases. Such interactions occur primarily in the major groove, which typically has a large, negative electrostatic potential (Chin et al. 1999; Klein et al. 2004). In the group II intron, cations are bound to helical motifs at a number of locations (M6–7, M16, M24, M30–31, K5, K10–11, K13, K15, K17, K20, K22, K28, and K31–32).
Among these sites, some of the K-sites bound to GU wobble pairs and GA mismatches exhibit particularly interesting properties. Ions K5, K11, K15, K17, K22 (see above), and K23 interact with GU wobble pairs. K5, K11, K17, and K23 adopt an arrangement identical to that of other monovalent ion-binding sites in 23S rRNA (Klein et al. 2004). For example, these ions are located in the plane of a GU wobble that is flanked by a GC pair and forming contact with O6 and O4 of the wobble nucleotides, and with O6 of the neighboring GC guanosine. In contrast, K15 and K22 bind GU wobble pairs that are not flanked by GC pairs and therefore adopt a different geometry, forming contact with only one of the wobble residues.
Finally, K6 binds in an unusual way to an imino GA mismatch in D2 (G270-A283), which is significant given the importance of GA motifs (Traub and Sussman 1982). More commonly, GA mismatches interact with multivalent metal ions (Heus et al. 1997; Rüdisser and Tinoco 2000; Villescas-Diaz and Zacharias 2003). For instance, sheared GA mismatches form inner-sphere contacts with partially dehydrated Mg2+ (Cate and Doudna 1996; Szep et al. 2003), and imino GA mismatches bind fully hydrated Mg2+ (Rüdisser and Tinoco 2000). In contrast, within the group II intron, the GA pair at K6 tolerates a diversity of different ions, including Tl+ (anomalous different Fourier density peak at 5.7σ), Rb+ (3.3σ), and Cs+ (4.9σ), which all form six inner-sphere contacts with A268, G269, and G270 and two interactions with water molecules from the bulk solvent. Ba2+ also binds to this site (4.7σ), albeit with different geometry (three inner-sphere contacts to G269, G270, and U271 and outer-sphere contacts with A268, G269, G270, and G284 mediated by a water molecule). In the presence of physiological ions (i.e., 4E8K, 4E8T, 4FAQ, 4FAW), K6 forms the same type of interactions as the heavy monovalent ions described above and it forms distances of 2.7–3.4 Å with its coordinating atoms, which is typical of potassium (Fig. 3). On the basis of these observations, we have chosen to interpret K6 as an unusual example of GA mismatch that has particular affinity for monovalent ions. However, the selectivity of G270-A283 for monovalent ions is not entirely clear-cut, because in one structure (4FAR) shorter distances (2.2–2.4 Å) compatible with magnesium binding were observed.
FIGURE 3.
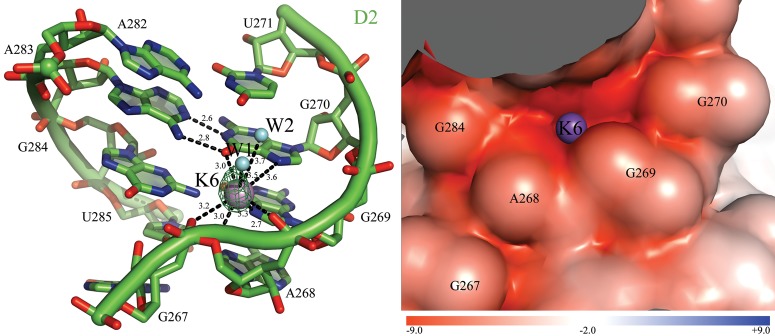
K6 binds an unusual GA imino mismatch in D2. (Left) Coordination of K6. The ion-binding site and the intron residues are colored as described in Figure 2. Inner-sphere coordination and hydrogen bonds are shown as black dashed lines for K6 and for the G270A283 imino mismatch and the corresponding distances are indicated in angstroms. Two water molecules completing the inner solvation sphere of K6 are also visible in the electron density and are represented here as cyan spheres (W1 and W2). Anomalous difference Fourier electron density map at the K6 site is shown as a green mesh at 4.0 σ for Tl+ (PDB id. 4E8Q). (Right) Electrostatic surface potential around K6. The color scale is indicated at the bottom in units of kT/e.
Metal ions involved in long-range tertiary interactions
Tertiary interaction motifs are crucial for the stabilization and specification of RNA tertiary structures (Butcher and Pyle 2011). Many of these motifs, such as the tetraloop–receptor interaction, require metal ions for stabilization or proper architectural organization, but a comprehensive analysis of metal requirements for most motifs has not been conducted. Thus, it is valuable to consider the metal ion occupancies and specificities of tertiary interaction motifs within the group II intron.
The five-way junction and the T-loop motif
The five-way junction is an elaborate motif at the center of D1 that establishes the location for all of the main D1 stems (D1A, D1B, D1C, D1d1, and D1(ii)), orienting them in the proper direction to form requisite long-range inter- and intradomain contacts. This motif contains a T-loop (D1A), a sharp backbone kink of 57° at position U26 (D1(ii)-D1A junction), a ribose zipper, and a type-I A-minor motif (D1(ii)-D1C interface) (Toor et al. 2010). The five-way junction region contains five M-sites (M11, M15, M20, M26, and M33) and two K-sites (K9 and K12).
The K12 ion (Fig. 4) lies just beneath A245 (D1d1), which is the base that intercalates into the T-loop (Toor et al. 2010). Binding of monovalent ions to this site of the T-loop may represent an idiosyncratic feature of group II introns, possibly related to the fact that this region is involved in crystal packing. Analogous structural sites in T-loops of different RNA structures (Krasilnikov and Mondragon 2003) possess affinity for multivalent metal ions. For instance, Pb2+ binds at an analogous position in the S-domain of bacterial ribonuclease P (Krasilnikov et al. 2003), while Mg2+ occupies a similar position in the 23S rRNA (Klein et al. 2001). Such comparisons indicate that T-loops may not be selective for ionic charge, but require site-bound metals in their region of maximal backbone curvature (Fig. 4).
FIGURE 4.
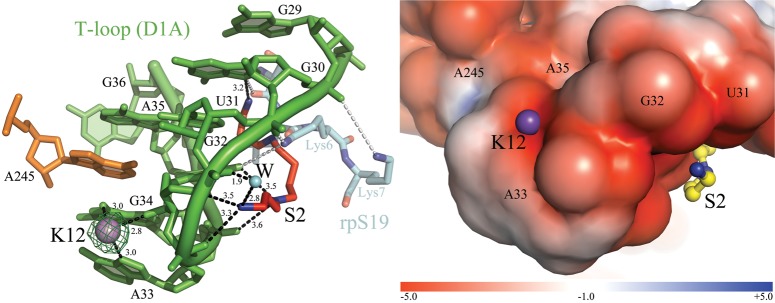
K12 and S2 bind the T-loop. (Left) The ion-binding sites and the intron residues are colored as described in Figure 2. Inner-sphere coordination bonds are shown as black dashed lines for K12 and S2 and the corresponding distances are indicated in angstroms. Anomalous difference Fourier electron density map at the K12 site is shown as a green mesh at 6.0 σ for Tl+ (PDB id. 4E8Q). The N-terminal polypeptide of ribosomal protein S19 (rpS19) is shown in semitransparent cyan sticks (PDB id. 1J5E). Gray dashed lines indicate contacts between the amino acids of rpS19 (Ser4-Lys6-Lys7) and nucleobases of 16S rRNA analogous to the intron T-loop (C1314-U1315-G1316, which correspond to G30-U31-G32, respectively). The 16S rRNA residues are not shown for clarity, but they superpose precisely over the intron T-loop residues (RMSD = 1.4 Å). (Right) Electrostatic surface potential around K12 and S2. The color scale is indicated at the bottom in units of kT/e.
The K9 and M33 ions bind close to the five-way junction, along the D1A stem. These ions are proximal to the tight kink in U26 mentioned above.
Finally, the ribose zipper that joins the base of D1C (U66 and A67) with the base of D1(ii) (U24) (Toor et al. 2010) contains two divalent ion sites, M15 and M20 (Supplemental Fig. S3). Interestingly, the latter forms a G-phosphate binding site near A67/G68 reminiscent of similar motifs found in 23S rRNA (Klein et al. 2004), but involving N7 of G68, rather than O6. Supported by its interactions with M15 and M20, the ribose zipper ejects A67 out of the D1C helix, enabling it to form a type I A-minor motif (Strobel 2002) with G248-C23 in D1(ii). At the same structural locus, the D1C strand opposite to A67 (A120/A121) comes close to G239 in D1d1 (ω), where D1C is effectively sandwiched between the D1(ii) and the D1d1 stems. M11 and M26 appear to glue the sandwich together (Supplemental Fig. S3), as M11 forms a magnesium clamp bridging the two SP oxygen atoms of residues C119 and G239 (D1d1, ω) and M26 is bound to the SP oxygen atom of A120.
The Z-anchor
The Z-anchor is a series of five nucleotides within the central loop of D1C (nucleotides 106–111) that form critical interactions with the catalytic center (Toor et al. 2008a). The Z-anchor includes the highly conserved interaction λ–λ′ (Boudvillain et al. 2000), which forms between A106 and G374 (D5), and ɛ–ɛ′ (Michel and Ferat 1995), which forms between G107 and U4 (5′-end). Tb3+-cleavage assays had previously predicted the presence of metals bound to the Z-anchor (Sigel et al. 2000). In agreement with that work, four ions can be identified at this position in our structures: M8, M13, K4 (see above and Fig. 2), and K25 (specific for Cs+). M8 and M13 bind the two SP phosphoryl oxygen atoms of G107 and A105, respectively. In this way, the ions may contribute to the stabilization of the 80° backbone kink formed by the two phosphate groups that flank A106. This backbone distortion projects the adenosine moiety of A106 (λ) toward D5 (λ′).
The θ–θ′ region
The θ–θ′ interaction (Costa et al. 1997) joins a tetraloop in D1C (G90 to A93) with a receptor in D2 (base pairs C272-G281 and U273-A280) through minor groove contacts (Supplemental Fig. S4). These residues form a minimized tetraloop-receptor motif, which lacks base stacking interactions and the AA-platform typical of other structures (Toor et al. 2010), but which surprisingly preserves the ion-binding pattern. In general, tetraloop-receptor motifs are observed to bind divalent and monovalent ions (Basu et al. 1998; Davis et al. 2007). In the group II intron, most ions near θ–θ′ are selective for monovalent cations, i.e., K7, K8, K14, and K16.
The most important ion of this set is K7, which binds the θ′ receptor in D2 (O4 of U273 and O6 of G274). K7 corresponds to potassium site 2 (P2/P8 tetraloop-receptor) in the structure of Azoarcus sp. BH72 group I intron (PDB id. 1U6B) (Adams et al. 2004) and to a potassium site near the L5b-J6a/6b motif in the structure of Tetrahymena thermophila ribozyme P4-P6 (Basu et al. 1998). This site can also be replaced by divalent ions, i.e., Mn2+ (Davis et al. 2007) or Ba2+ (this work, PDB id. 4E8V).
K14 and K16 also resemble tetraloop-bound ions in other RNA structures. K14 binds in a position identical to cobalt hexammine in group I intron tetraloops (Rüdisser and Tinoco 2000), coordinating phosphate oxygen atoms of the tetraloop and the base of a neighboring guanosine. Instead, K16, specific for Cs+ and Ba2+, binds the O6 atoms of two consecutive guanosines (G89 and G90), thus occupying a position identical to that of K+1017 in the P2/P8 tetraloop-receptor of Azoarcus sp. BH72 group I intron (PDB id. 1U6B) (Adams et al. 2004). The different sequences of the tetraloops in group I and II introns may be responsible for the different metal-binding selectivity.
The ω–ω′ ribose zipper and the EBS1 site
The ω–ω′ interaction is a ribose zipper between the first stem of D1d1 (ω) and a loop in D1d2 (ω′) (Toor et al. 2008a). This particular subclass of ribose zipper is specific to group IIC introns such as OiGIIi (Toor et al. 2008a). This motif positions exon binding site 1 (EBS1, located downstream ω′ in D1d2) near the five-way junction and consequently near the intron active site (Toor et al. 2008a). The ω–ω′ interaction and EBS1 bind six ions (M12, M18, M19, K15, K21, and K23; Supplemental Fig. S5).
The coordination and κ loops
The coordination and κ loops are two motifs in D1d1 that enable D1 to dock with D5 near the catalytic site and to align the two exon-binding sites (EBS1 and EBS3) for efficient exon ligation (Boudvillain and Pyle 1998; de Lencastre et al. 2005; Hamill and Pyle 2006). To ensure proper docking, the coordination and κ loops adopt highly unusual structures that involve multiple kinks (i.e., a backbone angle of 60° at U152) and narrow spaces between backbone chains (i.e., the phosphates of A134 and C145 are only 5.4 Å apart) (Toor et al. 2010). These regions were predicted to bind multivalent ions based on Tb3+, Mn2+, and Zn2+-induced cleavage assays (Sigel et al. 2000; Hertweck and Mueller 2001). Our structures confirm these predictions and reveal the presence of at least five M-sites: M3 (discussed above), M14, M21, M25, and M34 (Supplemental Fig. S1). Notably, one of the divalent ions near the κ loop (possibly M14 or M34) may correspond to the magnesium-binding site recently identified in an NMR study and shown to regulate the first step of group II intron folding (Donghi et al. 2013).
The S-turn
In D3, a canonical and highly conserved S-turn forms a flat platform that reinforces interactions with coaxially stacked elements of D1 and D2 (Pyle 2010). At the D3 S-turn, K19 is located ∼3.2–3.5 Å from neighboring atoms O6, N7, the RP phosphoryl oxygen of G320, N1 of A297, and O6 of G321 (Fig. 5), just upstream of the AA parallel interaction (A299-A319) (Leontis and Westhof 1998). Replacement with heavy ion analogs (Supplemental Table S1) reveals that K19 represents a highly dehydrated monovalent ion-binding site. Thus, K19 resembles an ion-binding site in the S-turn of the hairpin ribozyme (Walter et al. 2000; Ditzler et al. 2009). In the intron, K19 may enable G320 to stack with A268 near the base of D2 (Toor et al. 2010). K19 also interacts with G321, which is involved in stabilizing the toggled intermediate (Marcia and Pyle 2012). Given its location, K19 may play an important role in molding the architecture of D3, and its functional role in splicing should be investigated.
FIGURE 5.
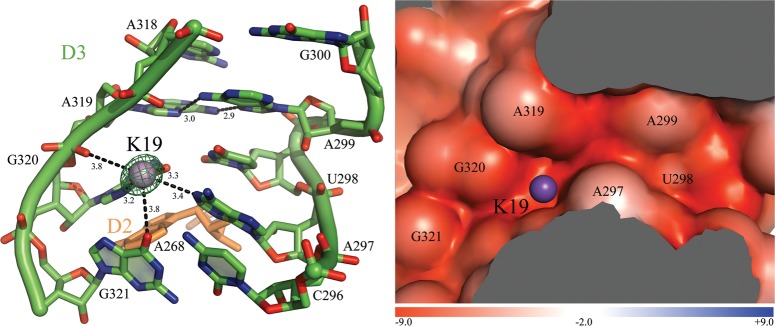
K19 binds the S-turn motif and the AA parallel interaction in D3. (Left) The ion-binding site and the intron residues are colored as described in Figure 2. Inner-sphere coordination and hydrogen bonds are shown as black dashed lines for K19 and for the A299A319 parallel interaction. The corresponding distances are indicated in angstroms. Anomalous difference Fourier electron density map at the K19 site is shown as a green mesh at 9.0 σ for Tl+ (PDB id. 4E8Q). (Right) Electrostatic surface potential around K19. The color scale is indicated at the bottom in units of kT/e.
Organic ion-binding sites
Polycation-binding sites
Two putative polycations (S1 and S2) were identified in proximity to long-range tertiary interactions involving D1A and D1B. S1 is proximal to the α–α′ interaction (Fig. 6), which is a phylogenetically conserved kissing loop between the α-loop sector (D1B) and the α′-loop (D1d2) (Michel et al. 1989). The α region adopts a sharp kink at its terminus, resulting in a backbone angle of 81° at phosphate G51 (Pyle 2010) and a concave cavity where S1 is located.
FIGURE 6.
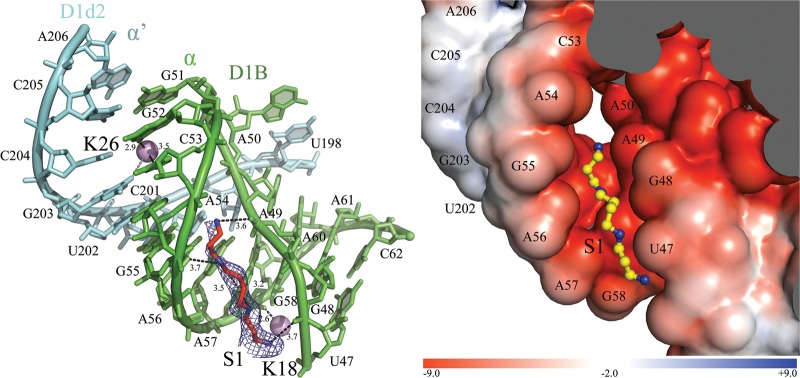
Ionic network at the α–α′ interaction. (Left) The ion-binding sites and the intron residues are colored as described in Figure 2 in the main text. The α–α′ interaction is indicated. Inner-sphere coordination and hydrogen bonds are shown as black dashed lines, with distances in angstroms. The interaction between K26 and O6 of G203 formed only in the presence of Cs+ is colored gray. The simulated annealing Fo–Fc electron density omit map is shown around the spermine molecule as a blue mesh at 3.0 σ. (Right) Electrostatic surface potential around S1. The color scale is indicated at the bottom in units of kT/e.
S2 is proximal to the T-loop, where it binds G32/A33/G34 along the extended major groove of the D1A stem, thus being located on the opposite side of K12 (Fig. 4; see above). Binding of amine groups to the face of the T-loop is interesting because this interaction pattern resembles the binding of basic amino acids to T-loops in certain ribonucleoproteins. For instance, the N-terminal polypeptide of ribosomal protein S19 binds similarly to a T-loop in 16S rRNA, establishing S2-like interactions (PDB id. 1J5E). Based on this similarity and on previous crosslinking experiments (Dai et al. 2008), the S sites may indicate positions of interaction between intron RNA and side-chains of encoded maturase proteins.
Zwitterionic ion-binding sites
Six putative sites (A1–6) compatible with the binding of HEPES molecules were also identified. At these positions, the RNA major groove displays patches of negative electrostatic potential, flanked by more positive regions (Supplemental Fig. S6). At these sites, low anomalous scattering signal can be detected in the Cs+ or Ba2+ structures, which is consistent with the presence of a heavy atom-like sulfur. The putative HEPES molecules, which are zwitterionic at the pH of the crystallization experiment (7.0), orient themselves such that their 2-hydroxyethyl-piperazine tails and their sulfonate head groups are positioned appropriately to bind the patches of negative and positive electrostatic surface potential along the RNA groove. As a result, the sulfur atom is located ∼3.8 Å from neighboring oxygen or nitrogen atoms of the RNA, as observed previously (Auffinger et al. 2004; Klein and Ferre-D'Amare 2006).
Specifically, the A-sites are located near consecutive Watson-Crick GC pairs (Fig. 1), which are known binding sites for sulfate ions (Auffinger et al. 2004) and sulfonic acid molecules (Klein and Ferre-D'Amare 2006). Thus, the A-sites resemble anionic-binding sites observed in other RNA structures, such as the Cricket Paralysis Virus intergenic region internal ribosome entry site domain 3 RNA, the P4–P6 domain of the Tetrahymena thermophila group I intron and the glmS ribozyme (Auffinger et al. 2004; Klein and Ferre-D'Amare 2006; Kieft et al. 2010). As anion-binders, consecutive GC pairs are known to possess affinity for the carboxylate groups of aspartate and glutamate in ribonucleoproteins (Kieft et al. 2010). Interestingly, the consecutive GC pairs in intron D1C are evolutionarily conserved and were identified as binding sites for the intron-encoded maturase, a protein that promotes intron folding and splicing (Matsuura et al. 2001; Dai et al. 2008). Therefore, while binding of zwitterions to RNA is typically of questionable physiological relevance, the A-sites in D1C of the O. iheyensis group II intron may represent sites of interaction with intron-encoded maturases. This hypothesis would be consistent with the colocalization of the A-sites in a region that is known to form an interface with protein in homologous introns from other organisms (Matsuura et al. 2001; Dai et al. 2008).
CONCLUSIONS
In conclusion, our comparison of group II intron structures shows that the intron interacts extensively and specifically with diverse ions. While the majority of ions are likely to stabilize the intron by binding weakly and in a delocalized manner, 74 ions bind tightly and with high occupancy, thus being visible in our crystal structures. Among these well-ordered ions, some suggest novel principles of RNA structural stabilization. For instance, we have described unusual ion binding properties of a GA mismatch, a T-loop, a tetraloop-receptor, and an S-turn. Interestingly, we have also revealed a role for organic ions that are bound to regions of the structure that had been previously predicted to interact with proteins. Finally and most importantly, we have extended our previous description of the catalytic heteronuclear metal center of the group II intron (Marcia and Pyle 2012), having identified and classified two additional groups of ions that have high occupancy and appear to support the architecture of the active site. These groups of ions may play important roles in positioning the active site and in aligning the reactants for catalysis.
Certainly, more questions remain and will be addressed by future biochemical and crystallographic work. The functional relevance of highly occupied and well-ordered ions observed in crystallographic structures must be proven by genetic and biochemical experiments. While the role in intron folding and splicing has already been established for some of the ions that we describe, our crystallographic investigations set the stage for a much broader functional investigation of many additional ions. Moreover, obtaining structures of the intron that include D6 would unveil the roles played by ions during the first step of splicing by transesterification and during the second step of splicing. Despite these open questions, the classification of the group II intron ion sites presented in this work, together with previous studies that mapped site-bound ions in RNAse P (Kazantsev et al. 2009), group I intron (Stahley et al. 2007), and ribosomal subunits (Klein et al. 2004), reveals the existence of common principles in RNA structural organization, which will be essential for determining and interpreting future RNA structures.
MATERIALS AND METHODS
Transcription, purification, crystallization, and structure determination
The constructs used in this work were cloned, transcribed in vitro, and purified in a native form, following protocols described previously (Toor et al. 2008a,b; Marcia and Pyle 2012). The structures were solved as described previously (Marcia and Pyle 2012; Marcia et al. 2013a). The positions of the anomalous scattering atoms were identified on the basis of anomalous difference Fourier electron density maps calculated using SFall and FFT in CCP4 (Ten Eyck 1973; Agarwal 1978). Electrostatic calculations were performed by solving a nonlinear Poisson-Boltzmann equation with APBS (Baker et al. 2001) as described previously (Kazantsev et al. 2009). Briefly, we treated the RNA as a low dielectric medium (ɛ = 2) (Misra and Draper 2001), and the surrounding solvent as a high dielectric continuum (ɛ = 78.5). We selected the volume enclosed by its water-accessible surface using a probe radius of 1.4 Å. We assigned atomic radii and charges with pdb2pqr (Dolinsky et al. 2004) according to AMBER force field parameters. We applied cubic B-spline discretization and nine-point harmonic averaging to the surface-based dielectric and ion-accessibility coefficients. Finally, we set Dirichlet boundary conditions using multiple Debye–Huckel functionality (Baker et al. 2001). The figures depicting the structures were drawn using PyMOL (DeLano 2009).
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article. A table listing all ion-binding sites and their corresponding occupancies, and figures representing ion binding at the coordination loop, at GU wobble pairs, at the 5-way junction, at the θ–θ′ and ω–ω′ interactions, and at conserved GC pairs, are provided as a single separate file.
ACKNOWLEDGMENTS
We thank the beamline scientists at 24-ID-C and E, NE-CAT, APS, for their thorough support during data collection. We also thank the referees of this paper for many helpful comments and suggestions. We acknowledge all members of the Pyle lab for valuable discussion and in particular Dr. Megan Fitzgerald and Dr. Olga Fedorova for critical reading of the manuscript. This project was supported by the National Institute of Health (RO1GM50313). A.M.P. is a Howard Hughes Medical Institute Investigator.
REFERENCES
- Abramovitz DL, Friedman RA, Pyle AM 1996. Catalytic role of 2′-hydroxyl groups within a group II intron active site. Science 271: 1410–1413 [DOI] [PubMed] [Google Scholar]
- Adams PL, Stahley MR, Kosek AB, Wang J, Strobel SA 2004. Crystal structure of a self-splicing group I intron with both exons. Nature 430: 45–50 [DOI] [PubMed] [Google Scholar]
- Agarwal RC 1978. A new least-squares refinement technique based on the fast Fourier transform algorithm. Acta Crystallogr A Found Crystallogr 34: 791–809 [Google Scholar]
- Auffinger P, Bielecki L, Westhof E 2004. Anion binding to nucleic acids. Structure 12: 379–388 [DOI] [PubMed] [Google Scholar]
- Auffinger P, Grover N, Westhof E 2011. Metal ion binding to RNA. Met Ions Life Sci 9: 1–35 [PubMed] [Google Scholar]
- Baker NA, Sept D, Joseph S, Holst MJ, McCammon JA 2001. Electrostatics of nanosystems: Application to microtubules and the ribosome. Proc Natl Acad Sci 98: 10037–10041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banatao DR, Altman RB, Klein TE 2003. Microenvironment analysis and identification of magnesium binding sites in RNA. Nucleic Acids Res 31: 4450–4460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basu S, Rambo RP, Strauss-Soukup J, Cate JH, Ferre-D'Amare AR, Strobel SA, Doudna JA 1998. A specific monovalent metal ion integral to the AA platform of the RNA tetraloop receptor. Nat Struct Biol 5: 986–992 [DOI] [PubMed] [Google Scholar]
- Boudvillain M, Pyle AM 1998. Defining functional groups, core structural features and inter-domain tertiary contacts essential for group II intron self-splicing: A NAIM analysis. EMBO J 17: 7091–7104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boudvillain M, de Lencastre A, Pyle AM 2000. A tertiary interaction that links active-site domains to the 5′ splice site of a group II intron. Nature 406: 315–318 [DOI] [PubMed] [Google Scholar]
- Boulanger SC, Belcher SM, Schmidt U, Dib-Hajj SD, Schmidt T, Perlman PS 1995. Studies of point mutants define three essential paired nucleotides in the domain 5 substructure of a group II intron. Mol Cell Biol 15: 4479–4488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butcher SE, Pyle AM 2011. The molecular interactions that stabilize RNA tertiary structure: RNA motifs, patterns, and networks. Acc Chem Res 44: 1302–1311 [DOI] [PubMed] [Google Scholar]
- Cate JH, Doudna JA 1996. Metal-binding sites in the major groove of a large ribozyme domain. Structure 4: 1221–1229 [DOI] [PubMed] [Google Scholar]
- Chin K, Sharp KA, Honig B, Pyle AM 1999. Calculating the electrostatic properties of RNA provides new insights into molecular interactions and function. Nat Struct Biol 6: 1055–1061 [DOI] [PubMed] [Google Scholar]
- Conn GL, Gittis AG, Lattman EE, Misra VK, Draper DE 2002. A compact RNA tertiary structure contains a buried backbone-K+ complex. J Mol Biol 318: 963–973 [DOI] [PubMed] [Google Scholar]
- Costa M, Deme E, Jacquier A, Michel F 1997. Multiple tertiary interactions involving domain II of group II self-splicing introns. J Mol Biol 267: 520–536 [DOI] [PubMed] [Google Scholar]
- Costa M, Michel F, Westhof E 2000. A three-dimensional perspective on exon binding by a group II self-splicing intron. EMBO J 19: 5007–5018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai L, Chai D, Gu SQ, Gabel J, Noskov SY, Blocker FJ, Lambowitz AM, Zimmerly S 2008. A three-dimensional model of a group II intron RNA and its interaction with the intron-encoded reverse transcriptase. Mol Cell 30: 472–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniels DL, Michels WJ Jr, Pyle AM 1996. Two competing pathways for self-splicing by group II introns: A quantitative analysis of in vitro reaction rates and products. J Mol Biol 256: 31–49 [DOI] [PubMed] [Google Scholar]
- Davis JH, Foster TR, Tonelli M, Butcher SE 2007. Role of metal ions in the tetraloop–receptor complex as analyzed by NMR. RNA 13: 76–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lencastre A, Pyle AM 2008. Three essential and conserved regions of the group II intron are proximal to the 5′-splice site. RNA 14: 11–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lencastre A, Hamill S, Pyle AM 2005. A single active-site region for a group II intron. Nat Struct Mol Biol 12: 626–627 [DOI] [PubMed] [Google Scholar]
- DeLano WL 2009. PyMOL molecular viewer: Updates and refinements. Abstr Pap Am Chem Soc 238 [Google Scholar]
- DeRose VJ 2003. Metal ion binding to catalytic RNA molecules. Curr Opin Struct Biol 13: 317–324 [DOI] [PubMed] [Google Scholar]
- Ditzler MA, Sponer J, Walter NG 2009. Molecular dynamics suggest multifunctionality of an adenine imino group in acid-base catalysis of the hairpin ribozyme. RNA 15: 560–575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolinsky TJ, Nielsen JE, McCammon JA, Baker NA 2004. PDB2PQR: An automated pipeline for the setup of Poisson–Boltzmann electrostatics calculations. Nucleic Acids Res 32: (Web Server issue): W665– W667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donghi D, Pechlaner M, Finazzo C, Knobloch B, Sigel RK 2013. The structural stabilization of the κ three-way junction by Mg(II) represents the first step in the folding of a group II intron. Nucleic Acids Res 41: 2489–2504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Draper DE 2004. A guide to ions and RNA structure. RNA 10: 335–343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Draper DE 2013. Folding of RNA tertiary structure: Linkages between backbone phosphates, ions, and water. Biopolymers 99: 1105–1113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ennifar E, Yusupov M, Walter P, Marquet R, Ehresmann B, Ehresmann C, Dumas P 1999. The crystal structure of the dimerization initiation site of genomic HIV-1 RNA reveals an extended duplex with two adenine bulges. Structure 7: 1439–1449 [DOI] [PubMed] [Google Scholar]
- Erat MC, Sigel RK 2007. Determination of the intrinsic affinities of multiple site-specific Mg2+ ions coordinated to domain 6 of a group II intron ribozyme. Inorg Chem 46: 11224–11234 [DOI] [PubMed] [Google Scholar]
- Erat MC, Sigel RK 2008. Divalent metal ions tune the self-splicing reaction of the yeast mitochondrial group II intron Sc.ai5γ. J Biol Inorg Chem 13: 1025–1036 [DOI] [PubMed] [Google Scholar]
- Feig AL, Uhlenbeck OC 1999. The role of metal ions in RNA biochemistry. In The RNA world (ed. Gesteland RF et al. ), pp. 287–320 Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York [Google Scholar]
- Freisinger E, Sigel RKO 2007. From nucleotides to ribozymes—a comparison of their metal ion binding properties. Coord Chem Rev 251: 1834–1851 [Google Scholar]
- Gordon PM, Piccirilli JA 2001. Metal ion coordination by the AGC triad in domain 5 contributes to group II intron catalysis. Nat Struct Biol 8: 893–898 [DOI] [PubMed] [Google Scholar]
- Hamill S, Pyle AM 2006. The receptor for branch-site docking within a group II intron active site. Mol Cell 23: 831–840 [DOI] [PubMed] [Google Scholar]
- Harding MM 2001. Geometry of metal-ligand interactions in proteins. Acta Crystallogr D Biol Crystallogr 57 (Pt 3): 401–411 [DOI] [PubMed] [Google Scholar]
- Harding MM 2002. Metal-ligand geometry relevant to proteins and in proteins: Sodium and potassium. Acta Crystallogr D Biol Crystallogr 58 (Pt 5): 872–874 [DOI] [PubMed] [Google Scholar]
- Hertweck M, Mueller MW 2001. Mapping divalent metal ion binding sites in a group II intron by Mn2+- and Zn2+-induced site-specific RNA cleavage. Eur J Biochem 268: 4610–4620 [DOI] [PubMed] [Google Scholar]
- Heus HA, Wijmenga SS, Hoppe H, Hilbers CW 1997. The detailed structure of tandem G.A mismatched base-pair motifs in RNA duplexes is context dependent. J Mol Biol 271: 147–158 [DOI] [PubMed] [Google Scholar]
- Holbrook SR, Sussman JL, Warrant RW, Kim SH 1978. Crystal structure of yeast phenylalanine transfer RNA. II. Structural features and functional implications. J Mol Biol 123: 631–660 [DOI] [PubMed] [Google Scholar]
- Jarrell KA, Peebles CL, Dietrich RC, Romiti SL, Perlman PS 1988. Group II intron self-splicing. Alternative reaction conditions yield novel products. J Biol Chem 263: 3432–3439 [PubMed] [Google Scholar]
- Kazantsev AV, Krivenko AA, Pace NR 2009. Mapping metal-binding sites in the catalytic domain of bacterial RNase P RNA. RNA 15: 266–276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieft JS, Chase E, Costantino DA, Golden BL 2010. Identification and characterization of anion binding sites in RNA. RNA 16: 1118–1123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein DJ, Ferre-D'Amare AR 2006. Structural basis of glmS ribozyme activation by glucosamine-6-phosphate. Science 313: 1752–1756 [DOI] [PubMed] [Google Scholar]
- Klein DJ, Schmeing TM, Moore PB, Steitz TA 2001. The kink-turn: A new RNA secondary structure motif. EMBO J 20: 4214–4221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein DJ, Moore PB, Steitz TA 2004. The contribution of metal ions to the structural stability of the large ribosomal subunit. RNA 10: 1366–1379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korolev N, Lyubartsev AP, Laaksonen A, Nordenskiold L 2002. On the competition between water, sodium ions, and spermine in binding to DNA: A molecular dynamics computer simulation study. Biophys J 82: 2860–2875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krasilnikov AS, Mondragon A 2003. On the occurrence of the T-loop RNA folding motif in large RNA molecules. RNA 9: 640–643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krasilnikov AS, Yang X, Pan T, Mondragon A 2003. Crystal structure of the specificity domain of ribonuclease P. Nature 421: 760–764 [DOI] [PubMed] [Google Scholar]
- Kruschel D, Sigel RK 2008. Divalent metal ions promote the formation of the 5′-splice site recognition complex in a self-splicing group II intron. J Inorg Biochem 102: 2147–2154 [DOI] [PubMed] [Google Scholar]
- Lambert D, Leipply D, Shiman R, Draper DE 2009. The influence of monovalent cation size on the stability of RNA tertiary structures. J Mol Biol 390: 791–804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leipply D, Lambert D, Draper DE 2009. Ion-RNA interactions thermodynamic analysis of the effects of mono- and divalent ions on RNA conformational equilibria. Methods Enzymol 469: 433–463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leontis NB, Westhof E 1998. A common motif organizes the structure of multi-helix loops in 16 S and 23 S ribosomal RNAs. J Mol Biol 283: 571–583 [DOI] [PubMed] [Google Scholar]
- Mahler J, Persson I 2012. A study of the hydration of the alkali metal ions in aqueous solution. Inorg Chem 51: 425–438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcia M, Pyle AM 2012. Visualizing group II intron catalysis through the stages of splicing. Cell 151: 497–507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcia M, Humphris-Narayanan E, Keating KS, Somarowthu S, Rajashankar K, Pyle AM 2013a. Solving nucleic acid structures by molecular replacement: Examples from group II intron studies. Acta Crystallogr D Biol Crystallogr 69 (Pt 11): 2174–2185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcia M, Somarowthu S, Pyle AM 2013b. Now on display: A gallery of group II intron structures at different stages of catalysis. Mob DNA 4: 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuura M, Noah JW, Lambowitz AM 2001. Mechanism of maturase-promoted group II intron splicing. EMBO J 20: 7259–7270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel F, Ferat JL 1995. Structure and activities of group II introns. Annu Rev Biochem 64: 435–461 [DOI] [PubMed] [Google Scholar]
- Michel F, Umesono K, Ozeki H 1989. Comparative and functional anatomy of group II catalytic introns—a review. Gene 82: 5–30 [DOI] [PubMed] [Google Scholar]
- Misra VK, Draper DE 2001. A thermodynamic framework for Mg2+ binding to RNA. Proc Natl Acad Sci 98: 12456–12461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pley HW, Flaherty KM, Mckay DB 1994. Three-dimensional structure of a hammerhead ribozyme. Nature 372: 68–74 [DOI] [PubMed] [Google Scholar]
- Podar M, Perlman PS, Padgett RA 1995. Stereochemical selectivity of group II intron splicing, reverse splicing, and hydrolysis reactions. Mol Cell Biol 15: 4466–4478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyle AM 2002. Metal ions in the structure and function of RNA. J Biol Inorg Chem 7: 679–690 [DOI] [PubMed] [Google Scholar]
- Pyle AM 2010. The tertiary structure of group II introns: Implications for biological function and evolution. Crit Rev Biochem Mol Biol 45: 215–232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyle AM, Fedorova O, Waldsich C 2007. Folding of group II introns: A model system for large, multidomain RNAs? Trends Biochem Sci 32: 138–145 [DOI] [PubMed] [Google Scholar]
- Quigley GJ, Teeter MM, Rich A 1978. Structural analysis of spermine and magnesium ion binding to yeast phenylalanine transfer RNA. Proc Natl Acad Sci 75: 64–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rüdisser S, Tinoco I Jr 2000. Solution structure of Cobalt(III)hexammine complexed to the GAAA tetraloop, and metal-ion binding to G.A mismatches. J Mol Biol 295: 1211–1223 [DOI] [PubMed] [Google Scholar]
- Schmidt U, Podar M, Stahl U, Perlman PS 1996. Mutations of the two-nucleotide bulge of D5 of a group II intron block splicing in vitro and in vivo: Phenotypes and suppressor mutations. RNA 2: 1161–1172 [PMC free article] [PubMed] [Google Scholar]
- Schnabl J, Suter P, Sigel RK 2012. MINAS—a database of Metal Ions in Nucleic AcidS. Nucleic Acids Res 40 (Database issue): D434–D438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigel RK, Vaidya A, Pyle AM 2000. Metal ion binding sites in a group II intron core. Nat Struct Biol 7: 1111–1116 [DOI] [PubMed] [Google Scholar]
- Stahley MR, Adams PL, Wang J, Strobel SA 2007. Structural metals in the group I intron: A ribozyme with a multiple metal ion core. J Mol Biol 372: 89–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefan LR, Zhang R, Levitan AG, Hendrix DK, Brenner SE, Holbrook SR 2006. MeRNA: A database of metal ion binding sites in RNA structures. Nucleic Acids Res 34 (Database issue): D131–D134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strobel SA 2002. Biochemical identification of A-minor motifs within RNA tertiary structure by interference analysis. Biochem Soc Trans 30 (Pt 6): 1126–1131 [DOI] [PubMed] [Google Scholar]
- Su LJ, Brenowitz M, Pyle AM 2003. An alternative route for the folding of large RNAs: Apparent two-state folding by a group II intron ribozyme. J Mol Biol 334: 639–652 [DOI] [PubMed] [Google Scholar]
- Su LJ, Waldsich C, Pyle AM 2005. An obligate intermediate along the slow folding pathway of a group II intron ribozyme. Nucleic Acids Res 33: 6674–6687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swisher JF, Su LJ, Brenowitz M, Anderson VE, Pyle AM 2002. Productive folding to the native state by a group II intron ribozyme. J Mol Biol 315: 297–310 [DOI] [PubMed] [Google Scholar]
- Szep S, Wang J, Moore PB 2003. The crystal structure of a 26-nucleotide RNA containing a hook-turn. RNA 9: 44–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ten Eyck LF 1973. Crystal physics, diffraction, theoretical and general crystallography. Acta Crystallogr A Found Crystallogr 29: 183–191 [Google Scholar]
- Toor N, Keating KS, Taylor SD, Pyle AM 2008a. Crystal structure of a self-spliced group II intron. Science 320: 77–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toor N, Rajashankar K, Keating KS, Pyle AM 2008b. Structural basis for exon recognition by a group II intron. Nat Struct Mol Biol 15: 1221–1222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toor N, Keating KS, Fedorova O, Rajashankar K, Wang J, Pyle AM 2010. Tertiary architecture of the Oceanobacillus iheyensis group II intron. RNA 16: 57–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traub W, Sussman JL 1982. Adenine-guanine base pairing ribosomal RNA. Nucleic Acids Res 10: 2701–2708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villescas-Diaz G, Zacharias M 2003. Sequence context dependence of tandem guanine:adenine mismatch conformations in RNA: A continuum solvent analysis. Biophys J 85: 416–425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waldsich C, Pyle AM 2008. A kinetic intermediate that regulates proper folding of a group II intron RNA. J Mol Biol 375: 572–580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter NG, Yang N, Burke JM 2000. Probing non-selective cation binding in the hairpin ribozyme with Tb(III). J Mol Biol 298: 539–555 [DOI] [PubMed] [Google Scholar]
- Yamada M, Miyawaki R, Nakai I, Izumi F, Nagashima K 1998. A Rietveld analysis of the crystal structure of ammonioleucite. Mineral J 20: 105–112 [Google Scholar]