Figure 1.
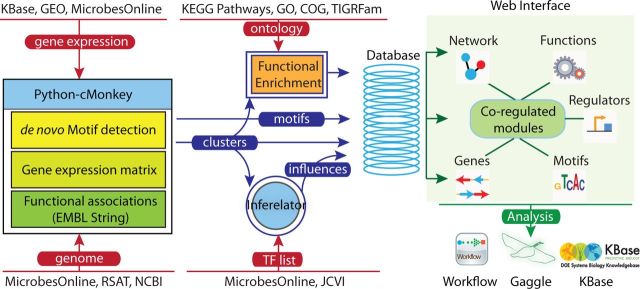
The network portal Framework. The network portal currently implements the Python–cMonkey algorithm for network inference. Publicly available gene-expression data and genomic information is collected from various databases along with functional associations from EMBL STRING. Conditionally co-regulated clusters of genes (modules) and motifs discovered by cMonkey are stored in the database. The most probable influences on these modules are identified by Inferelator, using a TF list collected from MicrobesOnline and JCVI-CMR. A Django-based Web interface dynamically creates module-centered views for Network, Functions, Genes, Regulators and Motifs. Further investigations of the networks can be performed by using interoperability and automation frameworks provided by Gaggle and Workflow, respectively.
