Abstract
Bacterial spores are a continuous problem for both food-based and health-related industries. Decades of scientific research dedicated towards understanding molecular and gene regulatory aspects of sporulation, spore germination and spore properties have resulted in a wealth of data and information. To facilitate obtaining a complete overview as well as new insights concerning this complex and tightly regulated process, we have developed a database-driven knowledge platform called SporeWeb (http://sporeweb.molgenrug.nl) that focuses on gene regulatory networks during sporulation in the Gram-positive bacterium Bacillus subtilis. Dynamic features allow the user to navigate through all stages of sporulation with review-like descriptions, schematic overviews on transcriptional regulation and detailed information on all regulators and the genes under their control. The Web site supports data acquisition on sporulation genes and their expression, regulon network interactions and direct links to other knowledge platforms or relevant literature. The information found on SporeWeb (including figures and tables) can and will be updated as new information becomes available in the literature. In this way, SporeWeb offers a novel, convenient and timely reference, an information source and a data acquisition tool that will aid in the general understanding of the dynamics of the complete sporulation cycle.
INTRODUCTION
During adverse environmental conditions, bacterial cells adopt developmental strategies, such as endospore formation, to ensure their survival. Bacterial spores are a continuous problem in food- and health-related industries because of their persistence after treatments and their ability to revert to vegetative cells through the process of germination (1,2). For instance, the disease of anthrax can persevere through the ingestion of spores that are able to survive the gastrointestinal tract and germinate to vegetative cells that produce lethal toxins (3). On the other hand, use of bacterial spores in the form of bioinsecticides (4), antigen delivery systems and vaccines (5) or probiotics (6,7) are upcoming fields that offer attractive applications. Therefore, a better understanding of the sporulation and germination processes, the level of heterogeneity therein, all genes and proteins involved, as well as influential effects of environmental factors have formed important fields of study for the past decades (8) and have provided a wealth of knowledge (9–12).
Most of the work on sporulation has been performed using the Gram-positive non-pathogenic organism Bacillus subtilis. The obtained data are extremely valuable and are often used as a reference model in sporulation research concerning other (pathogenic) bacteria, including Bacillus anthracis (13,14), Bacillus cereus (15,16) and various Clostridium species and strains that are of both medical and industrial importance (including Clostridium difficile, Clostridium perfringens and Clostridium botulinum) (17–22). This results in even more data and information, with various theories and speculations on molecular mechanisms, conservation of ‘core’ sporulation genes and emergence of evolutionary foundations (23,24). Sporulation of B. subtilis is an extremely complex cellular developmental process (11,25,26). New technological advances such as RNA sequencing, identification of small non-coding RNAs and increased understanding of processes through mathematical modelling allow us to answer questions beyond previous expectations (27–31), but simultaneously add to the complexity. Newly sequenced bacterial genomes of other spore-formers are increasingly available due to faster and cheaper methodologies and demand efficient analyses and readily available databases for comparison purposes (32). Therefore, a general overview on how spore formation is established (including which genes and regulatory pathways are involved) is very valuable to the field, but due to the complexity and dynamics increasingly difficult to obtain.
In this work, we describe a novel knowledge and data acquisition platform called SporeWeb (http://sporeweb.molgenrug.nl), which focuses on all developmental stages of sporulation of B. subtilis from a gene regulatory point of view. Through an interactive web interface querying the SporeWeb database (details available in Supplementary Material), it offers both a textual description and a graphical representation of the sequence of events throughout spore development (Figure 1). Importantly, it easily links to more catalogued information present on other knowledge Web sites, such as SubtiWiki (http://subtiwiki.uni-goettingen.de/) (33). This allows the reader to grasp what happens inside the cell on the regulatory level, with additional detailed information on key regulatory proteins involved. The database-driven SporeWeb Web site is dynamic and will be updated and extended when novel scientific data become available in the future. We believe that SporeWeb will be a continuous valuable asset to the research field of bacterial sporulation and will aid in our overall understanding of this complex developmental process.
Figure 1.
Graphical representation of the different levels in the SporeWeb Web site.
An interactive journey through all stages of the bacterial sporulation process
Commitment to sporulation is characterized by asymmetric cell division and expression of dedicated gene sets (34). This expression is tightly regulated in various sequential developmental stages and governed by complex biochemical communication between the two compartments of the cell (11). SporeWeb offers an interactive review of this complete process, which is the result of an extensive literature study. The homepage serves as a starting point for any sporulation stage of interest, which can be accessed by clicking on the homepage image or by selecting the ‘State’ in the menu bar. Subsequent pages offer both detailed descriptions and schematic representations of development. The figures are interactive and dynamic: they contain clickable items of interest and ensure updated information as genes are added to or edited in the database. Legends to the figures are described in the vertical grey bar on the right of the web page, whereas a review-like description of the sporulation state is shown below the figure, with direct links to relevant literature references.
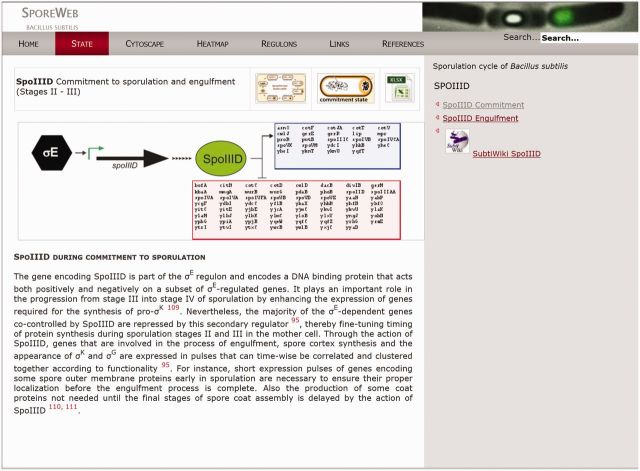
Sporulation-specific regulators and their regulons are described on individual pages, which can be accessed by clicking on the item in the schematic figures or via the menu bar. An example of such a page is shown in Figure 2. Genes under transcriptional control of a specific regulator have been indicated in blue or red boxes for transcriptional activation or repression, respectively. Additionally, there is a direct link to the SubtiWiki list of regulon members. Lists and descriptions of all genes within the regulon can be downloaded in the form of updatable Excel sheets by clicking on the coloured boxes or via the Excel icon in the top right corner (Figure 2).
Figure 2.
The ‘SpoIIID during commitment’ page on SporeWeb. Detailed information on the role of a sporulation-specific regulator during any stage in sporulation can be found on such pages. The schematic representation depicts its own regulation as well as the genes under its control; positively regulated in a blue box, negatively regulated in a red box. A detailed list containing all genes within the regulon can be accessed using the SubtiWiki link, or downloaded as an Excel file by clicking on the coloured boxes or the XLXS icon. To see what happens to this regulator during other stages in sporulation, simply click on the link provided in the grey bar on the right.
Graphic representations of regulon interactions define subgroups of co-regulated genes
As sporulation progresses, sporulation-specific sigma factors are expressed and activated in a spatial and temporal manner (11,26). Together with secondary regulator proteins, they control the timing, sequence and level of gene expression that are necessary for formation, maturation and release of the endospore. Various transcriptomic studies in B. subtilis have led to the identification of genes controlled by these sigma factors and other regulators to map the sporulation gene regulatory networks (35–42). In SporeWeb, we have visualized these networks using Cytoscape-generated layouts for every sporulation-specific stage (details available in Supplementary Material) (43). These layouts can be accessed via the ‘Cytoscape’ option in the menu bar, or the Cytoscape icon on the top of every State page.
An example of such a layout is shown in Figure 3A. This representation immediately shows which genes are under single, dual or even triple or quadruple control and which genes are co-regulated during a specific sporulation stage. There is a zoom-in function that allows the user to identify genes or regulators of interest. The name and direct links to the gene SubtiWiki and MicroScope MaGe pages (http://www.genoscope.cns.fr/agc/microscope/home/) are provided when the node is clicked. Furthermore, recently published spore-specific gene classification of the minimal sporulation gene set by Galperin et al. (24) has been integrated to immediately appreciate similarities and differences in regulation of functional classes of genes. Additionally, a visualization tool called ‘User Subset’ has been implemented in the Cytoscape menu bar that will allow the users to generate their own Cytoscape interaction figure of specific sporulation genes of interest (Figure 3B).
Figure 3.
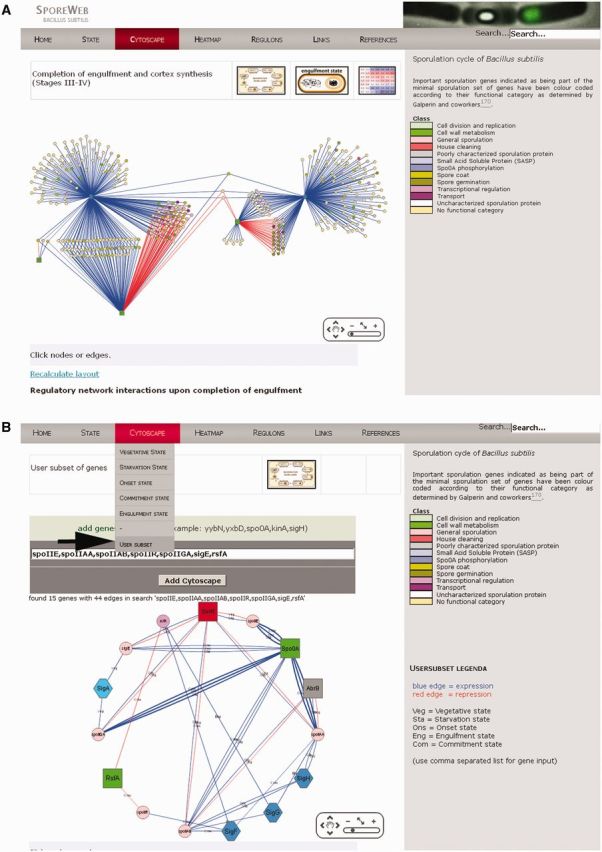
Cytoscape-generated layouts on gene regulatory networks. (A) Active regulators during completion of engulfment are indicated by green squares (proteins) or blue hexagons (sigma factors). Positive or negative effects on the transcription of genes (coloured circles) are indicated by connecting blue and red lines, respectively. Important sporulation genes indicated as being part of the minimal sporulation gene set have been colour-coded according to their functional category as determined by Galperin et al. (24). Genes unassigned to these functional categories are indicated as yellow circles. These layouts are available on SporeWeb for five different stages during spore formation. (B) A personal Cytoscape layout on specific genes of interest can be generated using the ‘User Subset’ option in the ‘Cytoscape’ menu (indicated by a black arrow). Genes (separated by a comma only) should be typed in the white bar and will be organized in a graphical network via the ‘Add Cytoscape’ option. Coloured nodes (shapes) and edges (lines) represent genes and connections as described for Figure 3A. Three-letter abbreviations at the edges indicate during which stage in sporulation the particular regulation is relevant. For Spo0A regulation, thicker edges represent high-threshold genes, whereas thin edges represent low-threshold genes (44).
Gene expression values during sporulation are visualized in informative heatmaps
A large group of important sporulation genes has already been identified and characterized, although many remain whose function and/or regulation is still unknown. To further visualize timing and co-regulation of gene expression during sporulation, we have displayed a recent sporulation-specific transcriptional dataset from Nicolas et al. (45) in colour-coded heatmaps. These heatmaps can be accessed via the ‘Heatmap' option on the menu bar, or via the heatmap icon at the top of every State page. Genes are categorized in classes based on their previously documented regulation. Their expression values during the complete course of sporulation as well as during three time points of germination are shown in colour-coded boxes (Figure 4). In this way, differences and similarities in expression between co-regulated genes are visible and can provide clues about possible function and/or regulation of uncharacterized genes. The heatmaps can be downloaded via the Excel icon on the top right of the page.
Figure 4.
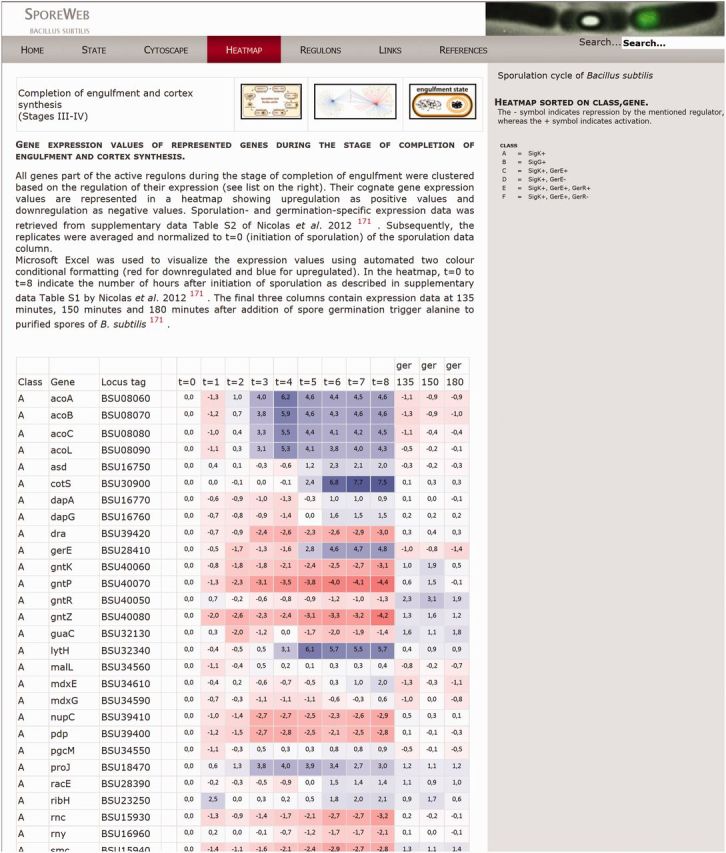
Heatmap representation of gene expression values during the engulfment state of sporulation. Tables like these are available on SporeWeb for five different stages in spore formation. Expression value data were derived from Nicolas et al. (45). Activation of gene expression is indicated as positive values in blue boxes, whereas downregulation is indicated as negative values in red boxes. Genes are categorized in classes listed A–L according to their documented regulation. Expression values are shown throughout the complete sporulation process (t = 0–t = 8) and for three time points taken during spore germination (ger).
Concluding remarks and perspectives
Knowledge on bacterial sporulation is rapidly growing, partly due to novel technological developments. This progress also reveals additional levels of complexity and makes it increasingly difficult to obtain a general understanding, especially for non-specialists in the field. Furthermore, rapid advances in DNA and RNA sequencing technologies have enabled faster and cheaper access to genomic- and transcriptional data of a large number of bacterial species. This leads to an expansion of our knowledge from laboratory-adapted model bacteria, such as B. subtilis, to more industrially or medically relevant species and strains. The data reveal high levels of conservation of certain genes or regulatory modules on the one hand and highlight important differences in gene presence/absence and regulatory events on the other, which have significant implications for the overall process of spore formation in specific groups of bacteria (23,24). The wealth of information that has been generated over decades by research on bacterial model organisms is extremely useful and usable as reference material for those bacterial species for which genetic manipulation and in vivo validation is less straightforward.
To provide researchers from all disciplines and expertise levels an accessible overview of the current state of knowledge on B. subtilis sporulation, we have constructed an interactive and graphical knowledge platform called SporeWeb. We believe that the potential of SporeWeb lies in the combination of a source of information on bacterial sporulation, an accessible starting point for further detailed investigation and a dynamic platform that can be adjusted and supplemented as new relevant data become available in the future. Moreover, the user-friendly interface and intuitive organization provide comprehensible data acquisition for both specialists and non-specialists in the field. Future perspectives for SporeWeb can include expansion to ‘sister’ Web sites. These can contain similar content on sporulation from a proteomics point of view or of (for instance) members of the B. cereus group (including B. anthracis and Bacillus thuringiensis) and/or Clostridium species, which would be a constructive addition to the applications of SporeWeb and a valuable contribution of knowledge to the entire field of sporulation research.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
TI Food and Nutrition, Wageningen, The Netherlands. Funding for open access charge: TI Food and Nutrition.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors are grateful for the time and dedication of Dr Adam Driks in thorough previewing of SporeWeb and they also thank many others in the microbiological and bacterial sporulation community who have come forward with useful suggestions and/or contributions.
REFERENCES
- 1.Eijlander RT, Abee T, Kuipers OP. Bacterial spores in food: how phenotypic variability complicates prediction of spore properties and bacterial behavior. Curr. Opin. Biotechnol. 2011;22:180–186. doi: 10.1016/j.copbio.2010.11.009. [DOI] [PubMed] [Google Scholar]
- 2.Augustin JC. Challenges in risk assessment and predictive microbiology of foodborne spore-forming bacteria. Food Microbiol. 2011;28:209–213. doi: 10.1016/j.fm.2010.05.003. [DOI] [PubMed] [Google Scholar]
- 3.Mock M, Fouet A. Anthrax. Annu. Rev. Microbiol. 2001;55:647–671. doi: 10.1146/annurev.micro.55.1.647. [DOI] [PubMed] [Google Scholar]
- 4.Schnepf E, Crickmore N, Van Rie J, Lereclus D, Baum J, Feitelson J, Zeigler DR, Dean DH. Bacillus thuringiensis and its pesticidal crystal proteins. Microbiol. Mol. Biol. Rev. 1998;62:775–806. doi: 10.1128/mmbr.62.3.775-806.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Amuguni H, Tzipori S. Bacillus subtilis: a temperature resistant and needle free delivery system of immunogens. Hum. Vaccin. Immunother. 2012;8:979–986. doi: 10.4161/hv.20694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bader J, Albin A, Stahl U. Spore-forming bacteria and their utilisation as probiotics. Benef. Microbes. 2012;3:67–75. doi: 10.3920/BM2011.0039. [DOI] [PubMed] [Google Scholar]
- 7.Permpoonpattana P, Hong HA, Khaneja R, Cutting SM. Evaluation of Bacillus subtilis strains as probiotics and their potential as a food ingredient. Benef. Microbes. 2012;3:127–135. doi: 10.3920/BM2012.0002. [DOI] [PubMed] [Google Scholar]
- 8.Gould GW. History of science–spores. J. Appl. Microbiol. 2006;101:507–513. doi: 10.1111/j.1365-2672.2006.02888.x. [DOI] [PubMed] [Google Scholar]
- 9.Higgins D, Dworkin J. Recent progress in Bacillus subtilis sporulation. FEMS Microbiol. Rev. 2012;36:131–148. doi: 10.1111/j.1574-6976.2011.00310.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Errington J. From spores to antibiotics via the cell cycle. Microbiology. 2010;156:1–13. doi: 10.1099/mic.0.035634-0. [DOI] [PubMed] [Google Scholar]
- 11.Hilbert DW, Piggot PJ. Compartmentalization of gene expression during Bacillus subtilis spore formation. Microbiol. Mol. Biol. Rev. 2004;68:234–262. doi: 10.1128/MMBR.68.2.234-262.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Moir A. How do spores germinate? J. Appl. Microbiol. 2006;101:526–530. doi: 10.1111/j.1365-2672.2006.02885.x. [DOI] [PubMed] [Google Scholar]
- 13.Liu H, Bergman NH, Thomason B, Shallom S, Hazen A, Crossno J, Rasko DA, Ravel J, Read TD, Peterson SN, et al. Formation and composition of the Bacillus anthracis endospore. J. Bacteriol. 2004;186:164–178. doi: 10.1128/JB.186.1.164-178.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fisher N, Hanna P. Characterization of Bacillus anthracis germinant receptors in vitro. J. Bacteriol. 2005;187:8055–8062. doi: 10.1128/JB.187.23.8055-8062.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.van der Voort M, García D, Moezelaar R, Abee T. Germinant receptor diversity and germination responses of four strains of the Bacillus cereus group. Int. J. Food Microbiol. 2010;139:108–115. doi: 10.1016/j.ijfoodmicro.2010.01.028. [DOI] [PubMed] [Google Scholar]
- 16.Vries de YP, Hornstra LM, de Vos WM, Abee T. Growth and sporulation of Bacillus cereus ATCC 14579 under defined conditions: temporal expression of genes for key sigma factors. Appl. Environ. Microbiol. 2004;70:2514–2519. doi: 10.1128/AEM.70.4.2514-2519.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xiao Y, Francke C, Abee T, Wells-Bennik MH. Clostridial spore germination versus bacilli: genome mining and current insights. Food Microbiol. 2011;28:266–274. doi: 10.1016/j.fm.2010.03.016. [DOI] [PubMed] [Google Scholar]
- 18.Paredes-Sabja D, Setlow P, Sarker MR. Germination of spores of Bacillales and Clostridiales species: mechanisms and proteins involved. Trends Microbiol. 2011;19:85–94. doi: 10.1016/j.tim.2010.10.004. [DOI] [PubMed] [Google Scholar]
- 19.Steiner E, Dago AE, Young DI, Heap JT, Minton NP, Hoch JA, Young M. Multiple orphan histidine kinases interact directly with Spo0A to control the initiation of endospore formation in Clostridium acetobutylicum. Mol. Microbiol. 2011;80:641–654. doi: 10.1111/j.1365-2958.2011.07608.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Burns DA, Minton NP. Sporulation studies in Clostridium difficile. J. Microbiol. Methods. 2011;87:133–138. doi: 10.1016/j.mimet.2011.07.017. [DOI] [PubMed] [Google Scholar]
- 21.Rosenbusch KE, Bakker D, Kuijper EJ, Smits WK. C. difficile 630Δerm Spo0A regulates sporulation, but does not contribute to toxin production, by direct high-affinity binding to target DNA. PLoS One. 2012;7:e48608. doi: 10.1371/journal.pone.0048608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Labbé RG, Dürre P. Sporulation of clostridia. In: Dürre P, editor. Handbook on Clostridia. FL, USA: CRC Press, Taylor & Francis Group; 2005. pp. 647–669. [Google Scholar]
- 23.de Hoon MJL, Eichenberger P, Vitkup D. Hierarchical evolution of the bacterial sporulation network. Curr. Biol. 2010;20:R735–R745. doi: 10.1016/j.cub.2010.06.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Galperin MY, Mekhedov SL, Puigbo P, Smirnov S, Wolf YI, Rigden DJ. Genomic determinants of sporulation in Bacilli and Clostridia: towards the minimal set of sporulation-specific genes. Environ. Microbiol. 2012;11:2870–2890. doi: 10.1111/j.1462-2920.2012.02841.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stragier P, Losick R. Molecular genetics of sporulation in Bacillus subtilis. Annu. Rev. Genet. 1996;30:297–241. doi: 10.1146/annurev.genet.30.1.297. [DOI] [PubMed] [Google Scholar]
- 26.Piggot PJ, Hilbert DW. Sporulation of Bacillus subtilis. Curr. Opin. Microbiol. 2004;7:579–586. doi: 10.1016/j.mib.2004.10.001. [DOI] [PubMed] [Google Scholar]
- 27.Silvaggi JM, Perkins JB, Losick R. Genes for small, noncoding RNAs under sporulation control in Bacillus subtilis. J. Bacteriol. 2006;188:532–541. doi: 10.1128/JB.188.2.532-541.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schmalisch M, Maiques E, Nikolov L, Camp AH, Chevreux B, Muffler A, Rodriguez S, Perkins J, Losick R. Small genes under sporulation control in the Bacillus subtilis genome. J. Bacteriol. 2010;192:5402–5412. doi: 10.1128/JB.00534-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liebal UW, Millat T, De Jong IG, Kuipers OP, Volker U, Wolkenhauer O. How mathematical modelling elucidates signalling in Bacillus subtilis. Mol. Microbiol. 2010;77:1083–1095. doi: 10.1111/j.1365-2958.2010.07283.x. [DOI] [PubMed] [Google Scholar]
- 30.Jabbari S, Heap JT, King JR. Mathematical modelling of the sporulation-initiation network in Bacillus subtilis revealing the dual role of the putative quorum-sensing signal molecule PhrA. Bull. Math. Biol. 2011;73:181–211. doi: 10.1007/s11538-010-9530-7. [DOI] [PubMed] [Google Scholar]
- 31.Levine JH, Fontes ME, Dworkin J, Elowitz MB. Pulsed feedback defers cellular differentiation. PLoS Biol. 2012;10:e1001252. doi: 10.1371/journal.pbio.1001252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rottger R, Rückert U, Taubert J, Baumbach J. How little do we actually know? – on the size of gene regulatory networks. IEEE/ACM Trans. Comput. Biol. Bioinform. 2012;9:1293–1300. doi: 10.1109/TCBB.2012.71. [DOI] [PubMed] [Google Scholar]
- 33.Mäder U, Schmeisky AG, Florez LA, Stülke J. SubtiWiki–a comprehensive community resource for the model organism Bacillus subtilis. Nucleic Acids Res. 2012;40:D1278–D1287. doi: 10.1093/nar/gkr923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Parker GF, Daniel RA, Errington J. Timing and genetic regulation of commitment to sporulation in Bacillus subtilis. Microbiology. 1996;142(Pt 12):3445–3452. doi: 10.1099/13500872-142-12-3445. [DOI] [PubMed] [Google Scholar]
- 35.Steil L, Serrano M, Henriques AO, Völker U. Genome-wide analysis of temporally regulated and compartment-specific gene expression in sporulating cells of Bacillus subtilis. Microbiology. 2005;151:399–420. doi: 10.1099/mic.0.27493-0. [DOI] [PubMed] [Google Scholar]
- 36.Cangiano G, Mazzone A, Baccigalupi L, Isticato R, Eichenberger P, De Felice M, Ricca E. Direct and indirect control of late sporulation genes by GerR of Bacillus subtilis. J. Bacteriol. 2010;192:3406–3413. doi: 10.1128/JB.00329-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Britton RA, Eichenberger P, Gonzalez-Pastor JE, Fawcett P, Monson R, Losick R, Grossman AD. Genome-wide analysis of the stationary-phase sigma factor (sigma-H) regulon of Bacillus subtilis. J. Bacteriol. 2002;184:4881–4890. doi: 10.1128/JB.184.17.4881-4890.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang ST, Setlow B, Conlon EM, Lyon JL, Imamura D, Sato T, Setlow P, Losick R, Eichenberger P. The forespore line of gene expression in Bacillus subtilis. J. Mol. Biol. 2006;358:16–37. doi: 10.1016/j.jmb.2006.01.059. [DOI] [PubMed] [Google Scholar]
- 39.Eichenberger P, Fujita M, Jensen ST, Conlon EM, Rudner DZ, Wang ST, Ferguson C, Haga K, Sato T, Liu JS, et al. The program of gene transcription for a single differentiating cell type during sporulation in Bacillus subtilis. PLoS Biol. 2004;2:e328. doi: 10.1371/journal.pbio.0020328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Eichenberger P, Jensen ST, Conlon EM, van Ooij C, Silvaggi J, Gonzalez-Pastor JE, Fujita M, Ben-Yehuda S, Stragier P, Liu JS, et al. The sigmaE regulon and the identification of additional sporulation genes in Bacillus subtilis. J. Mol. Biol. 2003;327:945–972. doi: 10.1016/s0022-2836(03)00205-5. [DOI] [PubMed] [Google Scholar]
- 41.Molle V, Fujita M, Jensen ST, Eichenberger P, Gonzalez-Pastor JE, Liu JS, Losick R. The Spo0A regulon of Bacillus subtilis. Mol. Microbiol. 2003;50:1683–1701. doi: 10.1046/j.1365-2958.2003.03818.x. [DOI] [PubMed] [Google Scholar]
- 42.Fawcett P, Eichenberger P, Losick R, Youngman P. The transcriptional profile of early to middle sporulation in Bacillus subtilis. Proc. Natl Acad. Sci. USA. 2000;97:8063–8068. doi: 10.1073/pnas.140209597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Saito R, Smoot ME, Ono K, Ruscheinski J, Wang PL, Lotia S, Pico AR, Bader GD, Ideker T. A travel guide to Cytoscape plugins. Nat. Methods. 2012;9:1069–1076. doi: 10.1038/nmeth.2212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fujita M, González-Pastor JE, Losick R. High- and low-threshold genes of the Spo0A regulon of Bacillus subtilis. J. Bacteriol. 2005;187:1357–1368. doi: 10.1128/JB.187.4.1357-1368.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nicolas P, Mäder U, Dervyn E, Rochat T, Leduc A, Pigeonneau N, Bidnenko E, Marchadier E, Hoebeke M, Aymerich S, et al. Condition-dependent transcriptome reveals high-level regulatory architecture in Bacillus subtilis. Science. 2012;335:1103–1106. doi: 10.1126/science.1206848. [DOI] [PubMed] [Google Scholar]