Abstract
Miniature inverted-repeat transposable elements (MITEs) are prevalent in eukaryotic species including plants. MITE families vary dramatically and usually cannot be identified based on homology. In this study, we de novo identified MITEs from 41 plant species, using computer programs MITE Digger, MITE-Hunter and/or Repetitive Sequence with Precise Boundaries (RSPB). MITEs were found in all, but one (Cyanidioschyzon merolae), species. Combined with the MITEs identified previously from the rice genome, >2.3 million sequences from 3527 MITE families were obtained from 41 plant species. In general, higher plants contain more MITEs than lower plants, with a few exceptions such as papaya, with only 538 elements. The largest number of MITEs is found in apple, with 237 302 MITE sequences. The number of MITE sequences in a genome is significantly correlated with genome size. A series of databases (plant MITE databases, P-MITE), available online at http://pmite.hzau.edu.cn/django/mite/, was constructed to host all MITE sequences from the 41 plant genomes. The databases are available for sequence similarity searches (BLASTN), and MITE sequences can be downloaded by family or by genome. The databases can be used to study the origin and amplification of MITEs, MITE-derived small RNAs and roles of MITEs on gene and genome evolution.
INTRODUCTION
Miniature inverted-repeat transposable elements (MITEs) are prevalent in eukaryotic genomes, and are believed to be deletion derivatives of DNA transposons (1,2). Like autonomous DNA transposons, MITEs usually have terminal inverted repeats (TIR), flanked by short direct repeats [also called target site duplication (TSD)]. Compared with autonomous DNA transposons, MITEs are often short (<800 bp) and do not encode transposases.
MITEs are often located in gene-rich euchromatic regions and are associated with genes (3,4). Several pieces of evidence suggest that MITEs may affect the expression of nearby genes. MITE Kiddo in rice was shown to upregulate the expression of Ubiquitin2 when inserted in its promoter region (5). However, in other cases, MITE insertions downregulate the expression of nearby genes (6,7). Such downregulation is most likely through small RNAs derived from MITE sequences (6,8). MITE transpositions generate much genetic diversity for a species (9–11). Considering the effects of MITEs on gene expression and variation of MITE insertions in different genotypes, MITEs may contribute to considerable phenotypic diversity as well (12).
The first MITE families were discovered through sequence analysis (i.e. identification of TIR and TSD sequences) of insertions of 100–600 bp (13,14). Recently, computer programs were developed to systematically identify MITEs from a database such as genome sequences (6,15–19). Among them, the most successful ones are MITE Digger, MITE-Hunter and RSPB, which identified the vast majority of MITEs in the sequenced genome of rice (6,18,19). The recently reported program MITE Digger is most efficient for de novo MITE identification, particularly in large genomes (19). RSPB is better at identifying MITE families with atypical structures such as MITEs with no TSD or short/diverse TIR sequences. Unfortunately, RSPB requires high computer capacity not found in most laboratories. We predicted that combining MITE Digger, MITE-Hunter and RSPB would allow the detection of a vast majority of, if not all, MITE families in a genome, with no prior information required. With the availability of the three MITE detecting programs and the genome sequences of many plant species, MITEs in several genomes can be readily identified and compared to further our understanding of MITE origin and evolution.
MITEs, as repetitive sequences, were included in other databases such as the The Institute for Genomic Research (TIGR) Plant Repeat Databases and Repbase (20,21). However, MITEs vary dramatically and usually cannot be identified through homology search between distantly related species, and consequently, only a small proportion of MITE families have been identified and included in these databases. In this study, MITEs were de novo identified from 41 plant species using computer programs MITE Digger, MITE-Hunter and/or RSPB. Each MITE family was annotated manually. All verified MITE families were stored in a database, P-MITE (for plant MITE). BLASTN search function was appended into the database. MITE sequences from each genome were downloadable. P-MITE will be helpful for the annotation of genes and genomic sequences. It can also be used to study the origin and amplification of MITEs, the comparative analysis between different species, the MITE-derived small RNAs and the roles of MITEs on gene and genome evolution, etc.
MATERIALS AND METHODS
Plant genomes used in this study
Forty-one sequenced and published genomes of plant species, including six lower plant species, were included in this study for MITE identification. The information of the 41 species and the Web sites for their genome sequences are listed in Supplementary Table S1. The MITEs from rice were identified and annotated in a previous study (6).
De novo identification of MITEs using MITE Digger, MITE-Hunter and RSPB
MITEs from 41 genomes were de novo identified using program MITE Digger, MITE-hunter and/or RSPB (6,18,19). First, program MITE-Hunter was used to run the sequences of each genome. The resulting groups of potential MITEs were manually checked for TSD and TIR sequences. Groups with no precise boundaries (terminals) or no TIR sequences were not considered as MITEs. The confirmed MITEs from MITE-Hunter were put into a database (MITE-Hunter database). To save running time, program RSPB was slightly modified so that the confirmed MITE sequences in the ‘MITE-Hunter database’ were skipped by RSPB. New groups of repetitive sequences with precise boundaries were reported and checked manually for TSDs and TIRs (Supplementary Figure S1). No TSD and TIR information is required to run RSPB, which identifies repetitive sequences with precise boundaries. In subsequent manual annotation, only repetitive sequences <800 bp and TSD/TIR features similar to known MITE superfamilies were maintained. Five species with large genomes or too many short contigs were not successful using RSPB. MITE Digger, released recently, was also used to run some genomes, including genomes >800 Mb. The statistics of MITE families identified in this study is shown in Supplementary Table S2. The number of MITE families that were detected by RSPB, but not by MITE Hunter, is shown in Supplementary Table S3.
Classification of MITE superfamily and family
A Perl script was written to cluster MITEs identified above into a family if they had significant sequence similarity (BLASTN e < 10−10) (6). MITE families were assigned into superfamilies based on their TIR and TSD sequences. Each MITE family in a genome was named as code_Abc#, where Ab is the first two letters from its genus name, c the first letter from its species name and # a consecutive number. Different superfamilies are represented by different codes, with DTT for Tc1/Mariner, DTM for Mutator, DTA for hAT, DTC for CACTA, DTH for PIF/Harbinger, DTP for P, DTN for Novosib and DTx for unknown (21–23). MITEs with ambiguous TSD and/or TIR features were annotated as unknown superfamily (DTx). MITE families preferentially inserted into simple tandem repeats (microsatellites) were considered as an independent group, MiM (MITEs inserted in microsatellite). A ‘representative’ element was chosen for each family, and the representative elements should have good TIR and perfect TSD sequences if possible. A MITE sequence was considered as a full-length element when its terminals were no more than 3 bp shorter than the representative sequence. To identify all MITE elements, including diverse and/or partial ones, in a genome, a library of all representative elements from each family was used as query sequences to search the entire genome sequence using RepeatMasker v3.2.9 (http://www.repeatmasker.org/).
RESULTS AND DISCUSSION
De novo identification of MITEs in 41 plant genomes
Program MITE-Hunter was applied to 41 plant genomes for genome-wide de novo identification of MITEs. RSPB was also used to run all but five genomes that are either >800 Mb or with too many contigs. MITE Digger was used to search some genomes, including four skipped by RSPB. The MITE sequences obtained from this study were used to execute a BLASTN search of the Repbase, the database most frequently used for repetitive sequences (21). More than 70% of MITE families identified from this study were not included in Repbase (< 10−10), MITE-Hunter, but not RSPB, due to too large genome. A total of 252 MITE families were obtained from maize, which include 97 novel families not covered by maize TE database. However, 61 MITE families listed in maize TE database were not identified by either MITE Digger or MITE-Hunter. The computing process of RSPB needs to be mended before it can be applied to large genomes, such as maize, to identify more novel MITE families.
The majority of MITEs were classified into five superfamilies, including Tc1/Mariner, PIF/Harbinger, CACTA, hAT and Mutator. Two superfamilies, P and Novosib, were detected in the genomes of lower plants, although they do not have Tc1/Mariner, CACTA and Mutator. Sixteen MITE families were unclassified owing to ambiguous TSD and/or TIR features. MiM is the least frequent in plant genomes (Supplementary Table S2). The MiM group is present in only 10 of the 41 genomes, with 41 893 elements from 33 families. The strawberry genome contains 14 MiM families, whereas the others have no more than four MiM families. Most elements of these MiM families, including the Micron family in rice (24), were inserted in (TA)n repeats, with only a few exceptions, in which they were inserted into (CA)n/(GT)n repeats. Elements from the MiM group have poor TIR sequences, and no conserved nucleotides were found in their terminals among different families. It remains unclear whether different MiM families belong to the same superfamily, i.e. activated by the same type of transposase. In contrast to the scarce MiM group, the Mutator superfamily has 852 390 elements in the 41 genomes included in this study, with an average of >20 790 elements per genome.
MITEs with significant nucleotide identities (BLASTN e < 10−10) were grouped into a family. The largest MITE family is the DTM_Mad25 from the apple genome, with 18 904 elements. The smallest MITE families, DTT_Sob24 and DTH_Sob33 from the Sorghum genome, have only one element.
The number of MITEs varies dramatically in different species. In general, the genomes of lower plants have relatively few MITEs (Table 1). No MITEs were detected in the genome of Cyanidioschyzon merolae using either MITE-Hunter or RSPB, and the genome of Selaginella moellendorffii harbors only 73 MITE elements. The number of MITEs also varies considerably among the genomes of higher plants. For example, only one MITE family with 538 elements was detected in the papaya genome, whereas 237 302 elements from 180 MITE families are present in the apple genome. Large variations in total number of MITE elements also occur between closely related species. For example, the Arabidopsis thaliana genome has only 3245 MITE elements, whereas its close relative, Arabidopsis lyrata, contains 18 039 MITE-related sequences. Similarly, the number of MITEs in the genome of watermelon (with 94 314 MITE elements) is seven times as much as in the genome of melon (with 12 991 MITE elements).
Table 1.
MITE in 41 plant genomes
| Species | Family | Genome size (Mb) | MITE |
|||
|---|---|---|---|---|---|---|
| Family number | Element number | Total length (Mb) | Percentage in genome | |||
| Phoenix dactylifera | Arecaceae | 381.56 | 33 | 39 990 | 8.22 | 2.15 |
| Arabidopsis thaliana | Brassicaceae | 119.67 | 43 | 3245 | 0.85 | 0.71 |
| Thellungiella parvula | Brassicaceae | 123.6 | 7 | 1161 | 0.32 | 0.26 |
| Arabidopsis lyrata | Brassicaceae | 206.67 | 121 | 18 039 | 4.64 | 2.24 |
| Thellungiella salsuginea | Brassicaceae | 208.87 | 54 | 5133 | 1.27 | 0.61 |
| Brassica rapa | Brassicaceae | 283.84 | 174 | 45 821 | 11.49 | 4.05 |
| Carica papaya | Caricaceae | 342.68 | 1 | 538 | 0.21 | 0.06 |
| Chlamydomonas reinhardtii | Chlamydomonadaceae | 111.1 | 20 | 3508 | 0.99 | 0.89 |
| Chlorella variabilis | Chlorellaceae | 46.16 | 2 | 83 | 0.04 | 0.08 |
| Cucumis sativus | Cucurbitaceae | 203.06 | 7 | 10 810 | 2.02 | 1.00 |
| Citrullus lanatus | Cucurbitaceae | 353.47 | 35 | 94 314 | 19.55 | 5.53 |
| Cucumis melo | Cucurbitaceae | 431.04 | 10 | 12 991 | 2.79 | 0.65 |
| Cyanidioschyzon merolae | Cyanidiaceae | 16.54 | 0 | 0 | 0.00 | 0.00 |
| Jatropha curcas | Euphorbiaceae | 297.67 | 17 | 18 975 | 4.81 | 1.61 |
| Ricinus communis | Euphorbiaceae | 350.63 | 33 | 13 205 | 3.24 | 0.93 |
| Manihot esculenta | Euphorbiaceae | 532.53 | 21 | 30 934 | 8.94 | 1.68 |
| Medicago truncatula | Fabaceae | 307.48 | 288 | 132 834 | 25.24 | 8.21 |
| Lotus japonicus | Fabaceae | 316.89 | 172 | 71 811 | 14.16 | 4.47 |
| Cajanus cajan | Fabaceae | 605.78 | 92 | 135 581 | 31.06 | 5.13 |
| Cannabis sativa | Fabaceae | 786.64 | 53 | 110 123 | 24.06 | 3.06 |
| Glycine max | Fabaceae | 973.34 | 126 | 169 379 | 27.69 | 2.84 |
| Physcomitrella patens | Funariaceae | 479.99 | 4 | 3718 | 0.58 | 0.12 |
| Linum usitatissimum | Linaceae | 318.25 | 28 | 14 409 | 3.51 | 1.10 |
| Theobroma cacao | Malvaceae | 327.35 | 13 | 10 364 | 3.45 | 1.06 |
| Musa acuminate | Musaceae | 472.96 | 9 | 15 835 | 2.22 | 0.47 |
| Coccomyxa subellipsoidea | Palmellaceae | 48.95 | 4 | 187 | 0.04 | 0.09 |
| Brachypodium distachyon | Poaceae | 271.92 | 222 | 83 272 | 12.86 | 4.73 |
| Oryza sativaa | Poaceae | 373.25 | 339 | 179 415 | 37.27 | 9.98 |
| Setaria italica | Poaceae | 405.78 | 178 | 69 264 | 15.60 | 3.85 |
| Sorghum bicolor | Poaceae | 738.58 | 275 | 112 307 | 29.63 | 4.01 |
| Zea mays | Poaceae | 2058.58 | 252 | 192 529 | 40.36 | 1.96 |
| Fragaria vesca | Rosaceae | 206.89 | 162 | 34 880 | 8.97 | 4.33 |
| Malus domestica | Rosaceae | 881.28 | 180 | 237 302 | 44.63 | 5.06 |
| Prunus persica | Rosaceae | 227.25 | 99 | 39 110 | 8.84 | 3.89 |
| Citrus sinensis | Rutaceae | 327.94 | 106 | 46 032 | 11.35 | 3.46 |
| Populus trichocarpa | Salicaceae | 417.14 | 22 | 35 081 | 7.49 | 1.80 |
| Selaginella moellendorffii | Selaginellaceae | 212.76 | 1 | 73 | 0.01 | 0.01 |
| Solanum lycopersicum | Solanaceae | 781.67 | 104 | 107 087 | 26.89 | 3.44 |
| Solanum tuberosum | Solanaceae | 797.83 | 171 | 170 392 | 38.65 | 4.84 |
| Vitis vinifera | Vitaceae | 486.19 | 35 | 61 065 | 14.69 | 3.02 |
| Volvox carteri | Volvocaceae | 131.16 | 14 | 2104 | 0.62 | 0.47 |
aThe MITE sequences from rice were retrieved from Lu et al. (25).
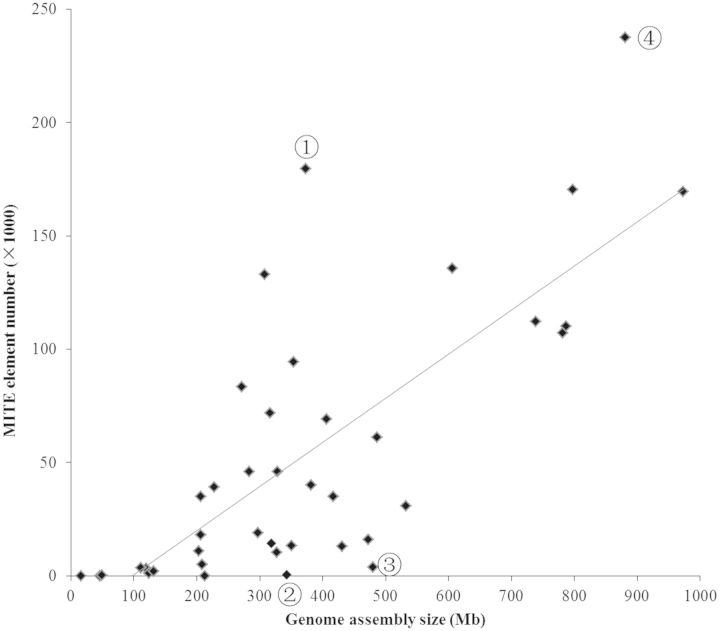
The number of MITEs in a genome is significantly correlated with its genome assembly size (r = 0.72, P < 0.01; Table 1; Figure 1). A similar correlation coefficient (r = 0.68, P < 0.01) was obtained when the six lower plants were excluded from the analysis. Nevertheless, several striking exceptions were observed. For example, the rice genome is only 373 Mb but has the third largest number (179 415) of MITEs among all species studied, whereas papaya with genome size (342 Mb) similar to that of rice, has only 538 elements of one MITE family (Table 1).
Figure 1.
Strong correlation between the number of MITEs and genome assembly size. Genomes with disproportionately low copy (➁ papaya and ➂ Physcomitrella patens) and high copy (➀ rice and ➃ apple) of MITEs are indicated.
The construction and the use of plant MITE database, P-MITE
A total of 2.3 million sequences of 3527 MITE families were obtained from 41 (including the rice genome) plant genomes. A series of databases containing MITEs from the 41 plant genomes was constructed. Elements from each of the 3527 MITE families were checked and annotated manually, and one element with better TSD and/or TIR features was chosen as a representative of the family. A database containing all representative elements was constructed, which can be used to study the structure of MITEs, such as their TSD and TIR features.
The aforementioned databases are collectively named as P-MITE (for plant MITE), and can be found in http://pmite.hzau.edu.cn/django/mite. The database is searchable using BLASTN algorithm. MITE sequences and representative elements can be downloaded by family or by genome.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online, including [26–66].
FUNDING
This work was supported by the ‘973’ National Key Basic Research Program [2009CB119000]; The National Natural Science Foundation of China (NSFC) [30921002]; and the Fundamental Research Funds for the Central Universities [2012ZYTS035]. Funding for open access charge: The Fundamental Research Funds for the Central Universities [2012ZYTS035].
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors are grateful to all the members in the Plant Genomics Lab, College of Horticulture and Forestry, Huazhong Agricultural University, for their manual annotation of MITE families.
REFERENCES
- 1.Feschotte C, Mouches C. Evidence that a family of miniature inverted-repeat transposable elements (MITEs) from the Arabidopsis thaliana genome has arisen from a pogo-like DNA transposon. Mol. Biol. Evol. 2000;17:730–737. doi: 10.1093/oxfordjournals.molbev.a026351. [DOI] [PubMed] [Google Scholar]
- 2.Feschotte C, Swamy L, Wessler SR. Genome-wide analysis of mariner-like transposable elements in rice reveals complex relationships with stowaway miniature inverted repeat transposable elements (MITEs) Genetics. 2003;163:747–758. doi: 10.1093/genetics/163.2.747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Han Y, Qin S, Wessler SR. Comparison of class 2 transposable elements at superfamily resolution reveals conserved and distinct features in cereal grass genomes. BMC Genomics. 2013;14:71. doi: 10.1186/1471-2164-14-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tu Z. Three novel families of miniature inverted-repeat transposable elements are associated with genes of the yellow fever mosquito, Aedes aegypti. Proc. Natl Acad. Sci. USA. 1997;94:7475–7480. doi: 10.1073/pnas.94.14.7475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yang G, Lee YH, Jiang Y, Shi X, Kertbundit S, Hall TC. A two-edged role for the transposable element Kiddo in the rice ubiquitin2 promoter. Plant Cell. 2005;17:1559–1568. doi: 10.1105/tpc.104.030528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lu C, Chen JJ, Zhang Y, Hu Q, Su WQ, Kuang HH. Miniature inverted-repeat transposable elements (MITEs) have been accumulated through amplification bursts and play important roles in gene expression and species diversity in Oryza sativa. Mol. Biol. Evol. 2012;29:1005–1017. doi: 10.1093/molbev/msr282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hollister JD, Gaut BS. Epigenetic silencing of transposable elements: a trade-off between reduced transposition and deleterious effects on neighboring gene expression. Genome Res. 2009;19:1419–1428. doi: 10.1101/gr.091678.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kuang H, Padmanabhan C, Li F, Kamei A, Bhaskar PB, Ouyang S, Jiang J, Buell CR, Baker B. Identification of miniature inverted-repeat transposable elements (MITEs) and biogenesis of their siRNAs in the Solanaceae: new functional implications for MITEs. Genome Res. 2009;19:42–56. doi: 10.1101/gr.078196.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Casa AM, Brouwer C, Nagel A, Wang L, Zhang Q, Kresovich S, Wessler SR. The MITE family heartbreaker (Hbr): molecular markers in maize. Proc. Natl Acad. Sci. USA. 2000;97:10083–10089. doi: 10.1073/pnas.97.18.10083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shirasawa K, Hirakawa H, Tabata S, Hasegawa M, Kiyoshima H, Suzuki S, Sasamoto S, Watanabe A, Fujishiro T, Isobe S. Characterization of active miniature inverted-repeat transposable elements in the peanut genome. Theor. Appl. Genet. 2012;124:1429–1438. doi: 10.1007/s00122-012-1798-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yaakov B, Ceylan E, Domb K, Kashkush K. Marker utility of miniature inverted-repeat transposable elements for wheat biodiversity and evolution. Theor. Appl. Genet. 2012;124:1365–1373. doi: 10.1007/s00122-012-1793-y. [DOI] [PubMed] [Google Scholar]
- 12.Chen J, Lu C, Zhang Y, Kuang H. Miniature inverted-repeat transposable elements (MITEs) in rice were originated and amplified predominantly after the divergence of Oryza and Brachypodium and contributed considerable diversity to the species. Mob. Genet. Elements. 2012;2:127–132. doi: 10.4161/mge.20773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bureau TE, Wessler SR. Tourist: a large family of small inverted repeat elements frequently associated with maize genes. Plant Cell. 1992;4:1283–1294. doi: 10.1105/tpc.4.10.1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wessler SR, Varagona MJ. Molecular basis of mutations at the waxy locus of maize: correlation with the fine structure genetic map. Proc. Natl Acad. Sci. USA. 1985;82:4177–4181. doi: 10.1073/pnas.82.12.4177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tu Z. Eight novel families of miniature inverted repeat transposable elements in the African malaria mosquito, Anopheles gambiae. Proc. Natl Acad. Sci. USA. 2001;98:1699–1704. doi: 10.1073/pnas.041593198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Santiago N, Herraiz C, Goni JR, Messeguer X, Casacuberta JM. Genome-wide analysis of the Emigrant family of MITEs of Arabidopsis thaliana. Mol. Biol. Evol. 2002;19:2285–2293. doi: 10.1093/oxfordjournals.molbev.a004052. [DOI] [PubMed] [Google Scholar]
- 17.Chen Y, Zhou F, Li G, Xu Y. MUST: a system for identification of miniature inverted-repeat transposable elements and applications to Anabaena variabilis and Haloquadratum walsbyi. Gene. 2009;436:1–7. doi: 10.1016/j.gene.2009.01.019. [DOI] [PubMed] [Google Scholar]
- 18.Han Y, Wessler SR. MITE-Hunter: a program for discovering miniature inverted-repeat transposable elements from genomic sequences. Nucleic Acids Res. 2010;38:e199. doi: 10.1093/nar/gkq862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang G. MITE Digger, an efficient and accurate algorithm for genome wide discovery of miniature inverted repeat transposable elements. BMC Bioinformatics. 2013;14:186. doi: 10.1186/1471-2105-14-186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ouyang S, Buell CR. The TIGR Plant Repeat Databases: a collective resource for the identification of repetitive sequences in plants. Nucleic Acids Res. 2004;32:D360–D363. doi: 10.1093/nar/gkh099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jurka J, Kapitonov VV, Pavlicek A, Klonowski P, Kohany O, Walichiewicz J. Repbase Update, a database of eukaryotic repetitive elements. Cytogenet. Genome Res. 2005;110:462–467. doi: 10.1159/000084979. [DOI] [PubMed] [Google Scholar]
- 22.Kapitonov VV, Jurka J. A universal classification of eukaryotic transposable elements implemented in Repbase. Nat. Rev. Genet. 2008;9:411–412. doi: 10.1038/nrg2165-c1. author reply 414. [DOI] [PubMed] [Google Scholar]
- 23.Wicker T, Sabot F, Hua-Van A, Bennetzen JL, Capy P, Chalhoub B, Flavell A, Leroy P, Morgante M, Panaud O, et al. A unified classification system for eukaryotic transposable elements. Nat. Rev. Genet. 2007;8:973–982. doi: 10.1038/nrg2165. [DOI] [PubMed] [Google Scholar]
- 24.Akagi H, Yokozeki Y, Inagaki A, Mori K, Fujimura T. Micron, a microsatellite-targeting transposable element in the rice genome. Mol. Genet. Genomics. 2001;266:471–480. doi: 10.1007/s004380100563. [DOI] [PubMed] [Google Scholar]
- 25.Lu C, Chen J, Zhang Y, Hu Q, Su W, Kuang H. Miniature inverted-repeat transposable elements (MITEs) have been accumulated through amplification bursts and play important roles in gene expression and species diversity in Oryza sativa. Mol. Biol. Evol. 2012;29:1005–1017. doi: 10.1093/molbev/msr282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Al-Dous EK, George B, Al-Mahmoud ME, Al-Jaber MY, Wang H, Salameh YM, Al-Azwani EK, Chaluvadi S, Pontaroli AC, DeBarry J, et al. De novo genome sequencing and comparative genomics of date palm (Phoenix dactylifera) Nat. Biotechnol. 2011;29:521–527. doi: 10.1038/nbt.1860. [DOI] [PubMed] [Google Scholar]
- 27.Hu TT, Pattyn P, Bakker EG, Cao J, Cheng JF, Clark RM, Fahlgren N, Fawcett JA, Grimwood J, Gundlach H, et al. The Arabidopsis lyrata genome sequence and the basis of rapid genome size change. Nat. Genet. 2011;43:476–481. doi: 10.1038/ng.807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Arabidopsis Genome Initiative. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature. 2000;408:796–815. doi: 10.1038/35048692. [DOI] [PubMed] [Google Scholar]
- 29.Wang X, Wang H, Wang J, Sun R, Wu J, Liu S, Bai Y, Mun JH, Bancroft I, Cheng F, et al. The genome of the mesopolyploid crop species Brassica rapa. Nat. Genet. 2011;43:1035–1039. doi: 10.1038/ng.919. [DOI] [PubMed] [Google Scholar]
- 30.Dassanayake M, Oh DH, Haas JS, Hernandez A, Hong H, Ali S, Yun DJ, Bressan RA, Zhu JK, Bohnert HJ, et al. The genome of the extremophile crucifer Thellungiella parvula. Nat. Genet. 2011;43:913–918. doi: 10.1038/ng.889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wu HJ, Zhang ZH, Wang JY, Oh DH, Dassanayake M, Liu BH, Huang QF, Sun HX, Xia R, Wu YR, et al. Insights into salt tolerance from the genome of Thellungiella salsuginea. Proc. Natl Acad. Sci. USA. 2012;109:12219–12224. doi: 10.1073/pnas.1209954109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ming R, Hou S, Feng Y, Yu Q, Dionne-Laporte A, Saw JH, Senin P, Wang W, Ly BV, Lewis KL, et al. The draft genome of the transgenic tropical fruit tree papaya (Carica papaya Linnaeus) Nature. 2008;452:991–996. doi: 10.1038/nature06856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Merchant SS, Prochnik SE, Vallon O, Harris EH, Karpowicz SJ, Witman GB, Terry A, Salamov A, Fritz-Laylin LK, Marechal-Drouard L, et al. The Chlamydomonas genome reveals the evolution of key animal and plant functions. Science. 2007;318:245–250. doi: 10.1126/science.1143609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Blanc G, Duncan G, Agarkova I, Borodovsky M, Gurnon J, Kuo A, Lindquist E, Lucas S, Pangilinan J, Polle J, et al. The Chlorella variabilis NC64A genome reveals adaptation to photosymbiosis, coevolution with viruses, and cryptic sex. Plant Cell. 2010;22:2943–2955. doi: 10.1105/tpc.110.076406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Guo S, Zhang J, Sun H, Salse J, Lucas WJ, Zhang H, Zheng Y, Mao L, Ren Y, Wang Z, et al. The draft genome of watermelon (Citrullus lanatus) and resequencing of 20 diverse accessions. Nat. Genet. 2013;45:51–58. doi: 10.1038/ng.2470. [DOI] [PubMed] [Google Scholar]
- 36.Garcia-Mas J, Benjak A, Sanseverino W, Bourgeois M, Mir G, Gonzalez VM, Henaff E, Camara F, Cozzuto L, Lowy E, et al. The genome of melon (Cucumis melo L.) Proc. Natl Acad. Sci. USA. 2012;109:11872–11877. doi: 10.1073/pnas.1205415109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Huang S, Li R, Zhang Z, Li L, Gu X, Fan W, Lucas WJ, Wang X, Xie B, Ni P, et al. The genome of the cucumber, Cucumis sativus L. Nat. Genet. 2009;41:1275–1281. doi: 10.1038/ng.475. [DOI] [PubMed] [Google Scholar]
- 38.Matsuzaki M, Misumi O, Shin IT, Maruyama S, Takahara M, Miyagishima SY, Mori T, Nishida K, Yagisawa F, Nishida K, et al. Genome sequence of the ultrasmall unicellular red alga Cyanidioschyzon merolae 10D. Nature. 2004;428:653–657. doi: 10.1038/nature02398. [DOI] [PubMed] [Google Scholar]
- 39.Sato S, Hirakawa H, Isobe S, Fukai E, Watanabe A, Kato M, Kawashima K, Minami C, Muraki A, Nakazaki N, et al. Sequence analysis of the genome of an oil-bearing tree, Jatropha curcas L. DNA Res. 2011;18:65–76. doi: 10.1093/dnares/dsq030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Prochnik S, Marri PR, Desany B, Rabinowicz PD, Kodira C, Mohiuddin M, Rodriguez F, Fauquet C, Tohme J, Harkins T, et al. The cassava genome: current progress, future directions. Trop. Plant Biol. 2012;5:88–94. doi: 10.1007/s12042-011-9088-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chan AP, Crabtree J, Zhao Q, Lorenzi H, Orvis J, Puiu D, Melake-Berhan A, Jones KM, Redman J, Chen G, et al. Draft genome sequence of the oilseed species Ricinus communis. Nat. Biotechnol. 2010;28:951–956. doi: 10.1038/nbt.1674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Varshney RK, Chen W, Li Y, Bharti AK, Saxena RK, Schlueter JA, Donoghue MT, Azam S, Fan G, Whaley AM, et al. Draft genome sequence of pigeonpea (Cajanus cajan), an orphan legume crop of resource-poor farmers. Nat. Biotechnol. 2012;30:83–89. doi: 10.1038/nbt.2022. [DOI] [PubMed] [Google Scholar]
- 43.van Bakel H, Stout JM, Cote AG, Tallon CM, Sharpe AG, Hughes TR, Page JE. The draft genome and transcriptome of Cannabis sativa. Genome Biol. 2011;12:R102. doi: 10.1186/gb-2011-12-10-r102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schmutz J, Cannon SB, Schlueter J, Ma J, Mitros T, Nelson W, Hyten DL, Song Q, Thelen JJ, Cheng J, et al. Genome sequence of the palaeopolyploid soybean. Nature. 2010;463:178–183. doi: 10.1038/nature08670. [DOI] [PubMed] [Google Scholar]
- 45.Sato S, Nakamura Y, Kaneko T, Asamizu E, Kato T, Nakao M, Sasamoto S, Watanabe A, Ono A, Kawashima K, et al. Genome structure of the legume, Lotus japonicus. DNA Res. 2008;15:227–239. doi: 10.1093/dnares/dsn008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Young ND, Debelle F, Oldroyd GED, Geurts R, Cannon SB, Udvardi MK, Benedito VA, Mayer KFX, Gouzy J, Schoof H, et al. The Medicago genome provides insight into the evolution of rhizobial symbioses. Nature. 2011;480:520–524. doi: 10.1038/nature10625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rensing SA, Lang D, Zimmer AD, Terry A, Salamov A, Shapiro H, Nishiyama T, Perroud PF, Lindquist EA, Kamisugi Y, et al. The Physcomitrella genome reveals evolutionary insights into the conquest of land by plants. Science. 2008;319:64–69. doi: 10.1126/science.1150646. [DOI] [PubMed] [Google Scholar]
- 48.Wang Z, Hobson N, Galindo L, Zhu S, Shi D, McDill J, Yang L, Hawkins S, Neutelings G, Datla R, et al. The genome of flax (Linum usitatissimum) assembled de novo from short shotgun sequence reads. Plant J. 2012;72:461–473. doi: 10.1111/j.1365-313X.2012.05093.x. [DOI] [PubMed] [Google Scholar]
- 49.Argout X, Salse J, Aury JM, Guiltinan MJ, Droc G, Gouzy J, Allegre M, Chaparro C, Legavre T, Maximova SN, et al. The genome of Theobroma cacao. Nat. Genet. 2011;43:101–108. doi: 10.1038/ng.736. [DOI] [PubMed] [Google Scholar]
- 50.D'Hont A, Denoeud F, Aury JM, Baurens FC, Carreel F, Garsmeur O, Noel B, Bocs S, Droc G, Rouard M, et al. The banana (Musa acuminata) genome and the evolution of monocotyledonous plants. Nature. 2012;488:213–217. doi: 10.1038/nature11241. [DOI] [PubMed] [Google Scholar]
- 51.Blanc G, Agarkova I, Grimwood J, Kuo A, Brueggeman A, Dunigan DD, Gurnon J, Ladunga I, Lindquist E, Lucas S, et al. The genome of the polar eukaryotic microalga Coccomyxa subellipsoidea reveals traits of cold adaptation. Genome Biol. 2012;13:R39. doi: 10.1186/gb-2012-13-5-r39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.International Brachypodium Initiative. Genome sequencing and analysis of the model grass Brachypodium distachyon. Nature. 2010;463:763–768. doi: 10.1038/nature08747. [DOI] [PubMed] [Google Scholar]
- 53.International Rice Genome Sequencing Project. The map-based sequence of the rice genome. Nature. 2005;436:793–800. doi: 10.1038/nature03895. [DOI] [PubMed] [Google Scholar]
- 54.Zhang G, Liu X, Quan Z, Cheng S, Xu X, Pan S, Xie M, Zeng P, Yue Z, Wang W, et al. Genome sequence of foxtail millet (Setaria italica) provides insights into grass evolution and biofuel potential. Nat. Biotechnol. 2012;30:549–554. doi: 10.1038/nbt.2195. [DOI] [PubMed] [Google Scholar]
- 55.Paterson AH, Bowers JE, Bruggmann R, Dubchak I, Grimwood J, Gundlach H, Haberer G, Hellsten U, Mitros T, Poliakov A, et al. The Sorghum bicolor genome and the diversification of grasses. Nature. 2009;457:551–556. doi: 10.1038/nature07723. [DOI] [PubMed] [Google Scholar]
- 56.Schnable PS, Ware D, Fulton RS, Stein JC, Wei F, Pasternak S, Liang C, Zhang J, Fulton L, Graves TA, et al. The B73 maize genome: complexity, diversity, and dynamics. Science. 2009;326:1112–1115. doi: 10.1126/science.1178534. [DOI] [PubMed] [Google Scholar]
- 57.Velasco R, Zharkikh A, Affourtit J, Dhingra A, Cestaro A, Kalyanaraman A, Fontana P, Bhatnagar SK, Troggio M, Pruss D, et al. The genome of the domesticated apple (Malus x domestica Borkh.) Nat. Genet. 2010;42:833–839. doi: 10.1038/ng.654. [DOI] [PubMed] [Google Scholar]
- 58.Shulaev V, Sargent DJ, Crowhurst RN, Mockler TC, Folkerts O, Delcher AL, Jaiswal P, Mockaitis K, Liston A, Mane SP, et al. The genome of woodland strawberry (Fragaria vesca) Nat. Genet. 2011;43:109–116. doi: 10.1038/ng.740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. International Peach Genome Initiative, Verde,I., Abbott,A.G., Scalabrin,S., Jung,S., Shu,S., Marroni,F., Zhebentyayeva,T., Dettori,M.T., Grimwood,J. et al. (2013) The high-quality draft genome of peach (Prunus persica) identifies unique patterns of genetic diversity, domestication and genome evolution. Nat. Genet., 45, 487–494. [DOI] [PubMed]
- 60.Xu Q, Chen LL, Ruan X, Chen D, Zhu A, Chen C, Bertrand D, Jiao WB, Hao BH, Lyon MP, et al. The draft genome of sweet orange (Citrus sinensis) Nat. Genet. 2013;45:59–66. doi: 10.1038/ng.2472. [DOI] [PubMed] [Google Scholar]
- 61.Tuskan GA, Difazio S, Jansson S, Bohlmann J, Grigoriev I, Hellsten U, Putnam N, Ralph S, Rombauts S, Salamov A, et al. The genome of black cottonwood, Populus trichocarpa (Torr. & Gray) Science. 2006;313:1596–1604. doi: 10.1126/science.1128691. [DOI] [PubMed] [Google Scholar]
- 62.Banks JA, Nishiyama T, Hasebe M, Bowman JL, Gribskov M, dePamphilis C, Albert VA, Aono N, Aoyama T, Ambrose BA, et al. The Selaginella genome identifies genetic changes associated with the evolution of vascular plants. Science. 2011;332:960–963. doi: 10.1126/science.1203810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tomato Genome Consortium. The tomato genome sequence provides insights into fleshy fruit evolution. Nature. 2012;485:635–641. doi: 10.1038/nature11119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Potato Genome Sequencing Consortium, Xu,X., Pan,S., Cheng,S., Zhang,B., Mu,D., Ni,P., Zhang,G., Yang,S., Li,R. et al. (2011) Genome sequence and analysis of the tuber crop potato. Nature, 475, 189–195. [DOI] [PubMed]
- 65.Jaillon O, Aury JM, Noel B, Policriti A, Clepet C, Casagrande A, Choisne N, Aubourg S, Vitulo N, Jubin C, et al. The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla. Nature. 2007;449:463–467. doi: 10.1038/nature06148. [DOI] [PubMed] [Google Scholar]
- 66.Prochnik SE, Umen J, Nedelcu AM, Hallmann A, Miller SM, Nishii I, Ferris P, Kuo A, Mitros T, Fritz-Laylin LK, et al. Genomic analysis of organismal complexity in the multicellular green alga Volvox carteri. Science. 2010;329:223–226. doi: 10.1126/science.1188800. [DOI] [PMC free article] [PubMed] [Google Scholar]