Abstract
‘Manually Curated Database of Rice Proteins’ (MCDRP) available at http://www.genomeindia.org/biocuration is a unique curated database based on published experimental data. Semantic integration of scientific data is essential to gain a higher level of understanding of biological systems. Since the majority of scientific data is available as published literature, text mining is an essential step before the data can be integrated and made available for computer-based search in various databases. However, text mining is a tedious exercise and thus, there is a large gap in the data available in curated databases and published literature. Moreover, data in an experiment can be perceived from several perspectives, which may not reflect in the text-based curation. In order to address such issues, we have demonstrated the feasibility of digitizing the experimental data itself by creating a database on rice proteins based on in-house developed data curation models. Using these models data of individual experiments have been digitized with the help of universal ontologies. Currently, the database has data for over 1800 rice proteins curated from >4000 different experiments of over 400 research articles. Since every aspect of the experiment such as gene name, plant type, tissue and developmental stage has been digitized, experimental data can be rapidly accessed and integrated.
INTRODUCTION
Decades of research on rice has generated several knowledge resources (1) such as genome sequences (2), annotations (3,4), transcript data (5,6), mutant resources (7,8) and germplasm collections (9,10) available through several excellent databases (4,6,7,11–16) or published literature, with a sole aim to understand every aspect of rice biology. Although, these databases have exhaustive data on rice, they do not precisely catalogue and integrate every detail of the experimental data published in scientific literature. Characterization of every genetic component in rice (17) requires seamless integration of data from various sources and thus, data curation has emerged as a major challenge (18,19). Bulk of scientific data are available as peer-reviewed articles accessible either through manual reading or through databases that mine the text of the articles (20,21). Extensive efforts have been done to develop efficient text-mining approaches (22–24) to enrich and integrate scientific data (25). Primarily, text-mining is done with a certain perspective and may not capture all the aspects of the data presented in the article. Moreover, text mining is a slow post-publication exercise, it is almost impossible to keep pace with the accumulation of published literature. Thus, there is a need to develop novel curation concepts which can deliver precise and fast integration of data (17). In the current study, we have demonstrated the feasibility of digitizing the experimental data instead of mining the text of the article. Digitization of experimental data itself offers several advantages:
Digitization would render the experimental data amenable to computerized search such that exact data from multiple articles can be searched/retrieved rapidly.
Digitization into a standard computer readable format would facilitate a seamless and semantic integration of data.
Data models for digitization of experimental data may be extended further to enable pre-publication integration of data. Thus, from the very beginning, the data would be in a format where it can be integrated into any database with minimal manual intervention. Such approach would help close the gap between the curated databases and the accumulated scientific literature.
Digitization of the experimental data via manual curation: concept
The manual curation workflow adopted in developing the database is designed to digitize the actual experimental data published in peer-reviewed articles. Such experimental data are mostly presented as images or graphs and is thus obviously not suitable for any computerized search. Moreover, there are no universal data representation standards for such experimental data. Thus, we developed the data curation workflow that was guided by the need to digitize experimental data as well as to lay foundation for development of standard representation of experimental data. The basic concept is depicted in Figure 1a and is dealt in detail elsewhere (Raghuvanshi et al. 2013, manuscript under preparation). In simple terms, every data point in a graph or image represents data of several dimensions such as gene name, plant type, tissue and growth conditions. Over the years, several ontologies and standard notations have been developed to represent at least some of these data types (26–28). Thus, a combined systematic use of these notations can represent information contained in every data point such that every data point is represented by a collection of these pre-defined terms. Since the notations would be alphanumeric in nature with pre-defined definition, the data can now be stored in any relational database and can be easily searched and correlated. Based on this concept, we have extensively used existing ontologies as well as in-house developed notations. To represent rice gene/protein, ‘Rice Genome Annotation Project (RGAP)’ locus ids have been primarily used (29). In cases where it was not possible to assign a RGAP id, the GenBank id has been used. Moreover, in several studies rice gene is studied in a heterologous plant system. In such cases, the experimental data may represent the expression of the heterologous gene. Thus, if the gene is from Arabidopsis, the ‘The Arabidopsis Information Resource’ (TAIR) (30) gene ids are used otherwise GenBank id of the heterologous gene has been used. The tissue and plant developmental stage is denoted by ‘Plant Ontology (PO)’ terms (31). Distinct PO terms have been used to represent plant developmental stage and plant tissue used in the analysis. In several cases, more than one PO term had to be used to represent a particular tissue such as ‘PO:0025034–PO:0005352–PO:0000074’ for ‘leaf–xylem–parenchyma’. The growth conditions such as temperature, water status, light or presence of any chemical such as hormones, metals, etc. have been denoted by ‘Environmental Ontology (EO)’ terms (27). In order to accurately denote environmental conditions, the ‘EO’ term is appended with additional data such as time of treatment, concentration, etc. The ‘Trait Ontology’ terms have been used to represent phenotypic or biochemical traits while the functional aspect of the protein is depicted with the help of ‘Gene Ontology’ (32). It may be noted that due to the vast complexity of the published experimental data, none of the above mentioned ontologies were sufficient to encode every aspect of data. Thus, a large number of new terms had to be defined for most ontologies to efficiently digitize the experimental data. The ids of all new ontology terms start with the numerical digit ‘1’ (such as TO:1000413—ascorbic acid content) to differentiate them from the pre-existing terms.
Figure 1.
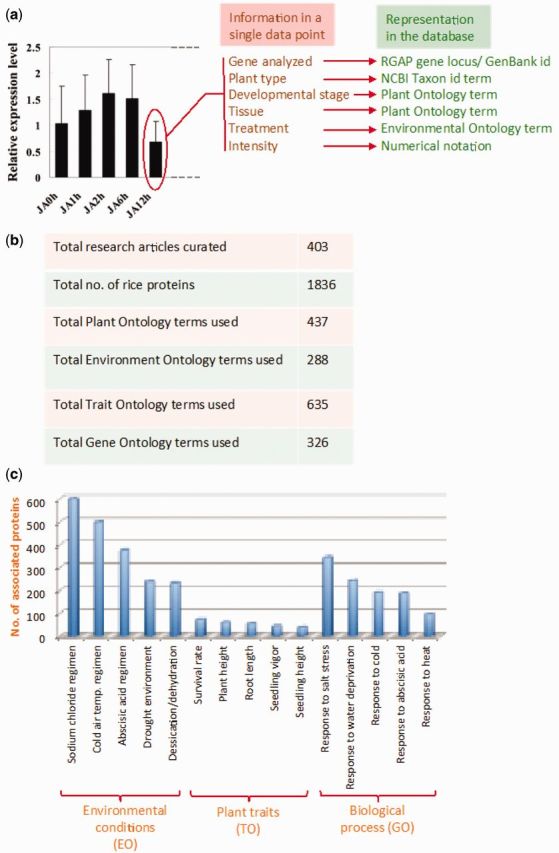
Details of the database. (a) Depiction of the concept for digitization of the experimental data. All aspects of the information in each data point (bars) is represented by an appropriate ontology/notation term. (b) Current status of the data curated in the database. (c) Distribution of the top five traits, environmental conditions and protein function in the curated database.
Besides the use of existing ontologies several other notations had to be developed to represent data for which no ontology exists. One of the most important is the notation to represent various experimental techniques. There are numerous different experimental techniques that are used in biological sciences and it is important to be aware of the experimental technique before any inference can be drawn from the data. Moreover, since all different type of experimental data is being coded with the help of similar set of ontologies and notations, it is very important to record the nature of the experiment. For example, a data point represented by ‘[LOC_Os05g46480/ PO:0009005 (roots)]’ would either represent the transcript levels of LOC_Os05g46480 in roots in a RT-PCR experiment or protein levels in a Western analysis. Thus, the context in which each term is used is essential for interpretation of the data. Notations were also defined to represent the type of rice plant that was used in the experiment, i.e. whether it was a wild type, transgenic over-expression, transgenic-RNAi line, mutant, etc. as well as to represent the promoters used to drive the transgene.
The current release of the database
The current release of the database catalogues data for over 1800 rice genes from ∼400 peer-reviewed research articles (Figure 1b). Data from more than 4000 different experiments have been digitized using in-house developed manual data curation models. These experiments are based on a total of ∼140 different experimental techniques mostly related to gene expression analysis, biochemical activity analysis, protein–protein interaction, DNA–protein interaction and cellular/sub-cellular localization. Majority of the curated articles are from the year 2007 onwards and published in a variety of peer-reviewed plant biology journals. Figure 1c shows the distribution of the ‘top five’ Environmental Ontology, Gene Ontology (biological process) and Trait Ontology terms. Based on the curated experimental data, these genes have been associated to more than 600 different traits and over 300 Gene Ontology terms. The most frequent traits include physical traits such as ‘survival rate’, ‘plant height’ and ‘root length’. Since there is a large collection of research articles on rice and the curation process is pretty extensive, we have tried to concentrate on studies related to abiotic stress in rice. This is clear from the distribution of the most frequent Gene Ontology terms that describe response to various abiotic stress conditions. The database also has protein–protein interaction data for 199 rice proteins as well as DNA–protein interaction data for 51 rice proteins.
How to access the database?
Every aspect (gene id, tissue, growth conditions, etc.) of the experimental data has been encoded with the help of defined notations (ontologies, etc.) and thus it is possible to retrieve the same set of data from several different perspectives. The database can be accessed either by browsing or searching for a particular keyword or term (Figure 2). A global overview of the data from different perspectives can be acquired by ‘browsing’ the database. Specific queries can be made by searching the database with the help of any of the ontology terms or with the help of a keyword. Both basic and advanced search options are available.
Figure 2.
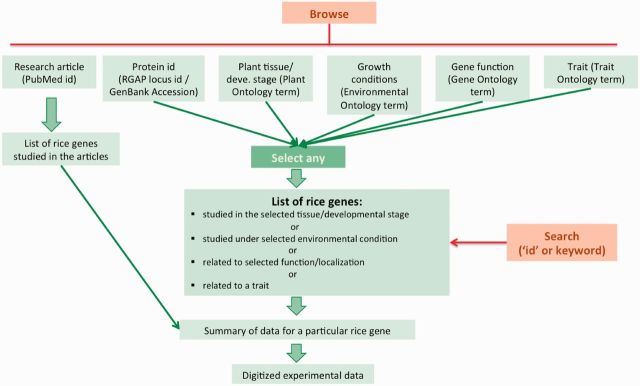
An overview of the different approaches to access the database. The database can be ‘Browsed’ from different perspectives such as PubMed id, gene locus id, plant tissue, environmental conditions, trait or protein function. Specific ‘Search’ can also be done by providing a keyword or an exact ontology term id.
Browsing
The entire content of the database is summarized from six different aspects for browsing. The data can be browsed by either PubMed id, rice gene locus id, plant tissue/developmental stage (Plant Ontology term), growth conditions (Environmental Ontology), phenotypic or biochemical trait (Trait Ontology) and gene function/localization (Gene Ontology term). While browsing by ‘PubMed id’, a list of all the articles curated in the database along with basic information such as title, authors, journal as well as the abstract is shown (Figure 3a). This list can be sorted either by PubMed id, journal or year of publication. A search option based on ‘PubMed id’, journal, author, year as well as any key word that may appear in the title of the article is also provided. On ‘clicking’ the PubMed id of interest, a list of all the rice gene loci that have been studied in that article is shown followed by a list of experiments that have been digitized (Figure 3b). Further, selecting any rice gene locus opens the ‘Rice gene details’ page that summarizes the information about the rice gene as presented in the selected article (Figure 4). Initially, the data shown would be restricted to the selected article, however, it is also possible to access data for the rice gene locus gathered from all the articles that have been curated in the database by selecting the option ‘Search entire database for LOC … .’. The information in the ‘Rice gene details’ page is divided into several sections. The section titled ‘Basic information’ presents the information about the protein domain as available in the Pfam database (33). The ‘Functional details’ sections list all the GO terms that have been mapped to the rice gene based on the experimental data. ‘Clicking’ on any of the GO term id provides the details of the experiment on the basis of which the GO term has been assigned. GO terms with (RGAP) tag have been acquired from the RGAP database (30). Similarly, the ‘Plant developmental stage/tissue details’ section lists all the tissue or developmental stages where the selected rice gene has been studied. The presence or absence of expression/protein activity in a particular tissue/developmental stage is indicated by ‘(+)’ or ‘(–)’ sign, respectively. Thus, ‘(–)’ means that although expression level or gene activity of the gene has been studied in that particular tissue but no detectable expression or activity was found. Selecting any of the PO terms presents the digitized data of the related experiment. In most cases, the data pertaining to a gene in a particular PO would only be part of the total experiment (e.g. one bar of the entire real-time RT-PCR graph). In such cases, initially data for only the selected data point would be shown, however, one can see the data for the entire experiment by selecting the ‘Show data for entire experiment’ option. The information about the environmental conditions under which the expression and activity of the gene has been studied is given as a list of Environmental Ontology terms in the ‘Environment details’ section. Thus, for example the affect of abiotic stress conditions such as drought, heat, salinity or hormones and other chemicals is recorded with the help of the EO terms. Selecting any particular EO term shows the digitized experimental data. Again ‘Show data for entire experiment’ option can be used if one wants to see the data of the entire experiment. The section entitled ‘Trait details’ summarizes all the phenotypic or biochemical traits that have been associated with the selected gene. The digitized experimental details can be accessed as explained above in the case of ‘Plant developmental stage/tissue details’. The last section summarizes the physical interaction data (protein–protein and protein–DNA) for the rice protein.
Figure 3.
Screenshot of the ‘Browse by PubMed id’ option. Initially a list of all the curated research articles would be shown along with data such as PubMed id, year of publication, journal, authors, title, abstract (a). Selecting any of the PubMed ids would give a list of all the rice proteins studied in the selected article (b).
Figure 4.
Screenshot of page showing details of a particular rice gene locus. The data for a particular gene locus from all the articles curated in the database is summarized under different heads. Part of the page (central) has been removed to accommodate the figure within a page.
The database can also be browsed through rice gene ids. This gives a list of all the rice genes that have been curated in the database. The first column of the table contains rice gene id that has been curated which in most cases is the RGAP locus id. A significant number of papers do not cite the relevant RGAP locus id. In such cases, RGAP locus id was assigned on the basis of sequence alignment at DNA level. If there was 100% identity and coverage between the gene sequence (mostly GenBank id) mentioned in the article and any RGAP locus id then the data were encoded using RGAP locus id. In case of lower but significant similarity the data were encoded using the original id (or the corresponding protein id, in case the mentioned id is of a genomic sequence or cDNA) as mentioned in the article and the closest RGAP locus id is mentioned in the column titled ‘Nearest RGAP protein id’. The column ‘Associated proteins’ gives information of all the rice genes that have been experimentally shown to be associated with the concerned rice protein. Rice proteins having physical association are indicated with tag ‘(I)’ whereas proteins that may affect the expression are indicated by tag ‘(R)’. If a protein affects the function of the concerned protein then it is indicated by tag ‘(F)’. Selecting any gene id in the first column opens the ‘Rice gene details’ which has been described as above.
Similarly, the database can also be browsed by Plant Ontology term. This gives the list of all the Plant Ontology terms that have been curated in the database. Selecting any one term lists all the rice genes that have been studied in the selected plant tissue or developmental stage. Further selecting the rice gene id opens the ‘Rice gene details’ page. Browsing by Environmental Ontology, Trait Ontology and Gene Ontology follows the similar trend.
Searching the database
The database can be searched on the basis of gene id, plant developmental stage, plant part/tissue, environmental conditions, associated traits, associated molecular function, associated biological process or cellular localization. The search can be done by either specifying the exact ‘term’ id or the keyword. In order to facilitate this the search function operates in two tiers. If an exact ‘term id’ is defined (such as protein id, ontology term id) the relevant results are shown immediately. In case a keyword is given, in the first stage a list of all the related terms is displayed. The user may then select one or more of these terms and then proceed to the second stage where data relevant to the selected term is shown. It may be noted that search would be much faster when an exact id is specified such as ‘PO:OOO9005’ instead of the keyword ‘root’. It is also possible to formulate search with multiple terms and boolean operators AND and OR. The output of the search is a list of rice gene ids as well as the link to the exact experiment (PubMed id and experiment no.) where the search term has been used.
Major highlights
Digitization of experimental data of published peer-reviewed studies is a relatively less explored dimension of data curation. Nevertheless, it offers several advantages. Some of the major aspects that stand out are as follows:
Seamless semantic integration of data: The digitization of the experimental data has been done using the same basic elements (i.e. ontologies and notations) thus providing a natural connectivity of data coming from different experimental set-ups.
Universality of the basic concept: The fundamental models developed to digitize the experimental data on rice genes are universal in nature and can be easily adopted for other organisms as well. The basic requirement is the availability of organism specific ontologies. The current exercise has led to a significant enrichment of plant ontologies as a large number of novel terms had to be defined in order to digitize the published experimental data.
In-depth and precise functional annotation: One of the major advantages of indexing the experimental data itself is the preciseness of the functional annotation of the genes. The assigned GO terms lie very low in the hierarchy and are thus more precise in nature. There is a direct connection of the associated ‘GO’ term with the experimental data that was used to assign the GO term. Another aspect is the cataloguing of the dependencies of a particular GO term assigned to the protein. For example a ‘kinase’ activity of the protein may be dependent on the presence of other protein, certain environmental conditions or restricted to a plant tissue. Such dependencies have also been catalogued.
Learning resource: Such databases can be great learning resources for young researchers. Naïve questions such as ‘What concentration of a particular chemical (hormone/metal ion/drug, etc.) should be used while designing experiments?’ A simple search with a suitable ‘EO’ term would line up all the experiments conducted with the concerned chemical in a matter of seconds.
Future perspective
The database will be updated every 6 months and as the curation progresses the data would be enriched by inclusion of studies from diverse aspects such as biotic stress, yield, etc. The ultimate aim is to digitize all the relevant articles but since the task is enormous we are proceeding in a methodical manner so that important aspects such stress biology (abiotic or biotic), yield are better represented. The concept of digitization of the experimental data from peer-reviewed research articles is relatively new and is bound to evolve. Better user-friendly portals are being developed to facilitate easy and fast digitization of experimental data. Such portals would also enable active participation of third party curators to meet the challenge of curating all the articles. Lab data management implementation may integrate these curation models so that they can be used not only to store data but also in designing of the experiment from the very beginning. Thus, at the time of publication, the data would already be in a format compatible for integration in the database.
FUNDING
Funding for open access charge: The Department of Biotechnology, Government of India, India. The work is supported by grant from The Department of Biotechnology, Government of India, India.
Conflict of interest statement. None declared.
REFERENCES
- 1.Yang Y, Li Y, Wu C. Genomic resources for functional analyses of the rice genome. Curr. Opin. Plant Biol. 2013;16:157–163. doi: 10.1016/j.pbi.2013.03.010. [DOI] [PubMed] [Google Scholar]
- 2.IRGSP. The map-based sequence of the rice genome. Nature. 2005;436:793–800. doi: 10.1038/nature03895. [DOI] [PubMed] [Google Scholar]
- 3.Itoh T, Tanaka T, Barrero RA, Yamasaki C, Fujii Y, Hilton PB, Antonio BA, Aono H, Apweiler R, Bruskiewich R, et al. Curated genome annotation of Oryza sativa ssp. japonica and comparative genome analysis with Arabidopsis thaliana. Genome Res. 2007;17:175–183. doi: 10.1101/gr.5509507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sakai H, Lee SS, Tanaka T, Numa H, Kim J, Kawahara Y, Wakimoto H, Yang C, Iwamoto M, Abe T, et al. Rice Annotation Project Database (RAP-DB): an integrative and interactive database for rice genomics. Plant Cell Physiol. 2013;54:e6. doi: 10.1093/pcp/pcs183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kikuchi S, Satoh K, Nagata T, Kawagashira N, Doi K, Kishimoto N, Yazaki J, Ishikawa M, Yamada H, Ooka H, et al. Collection, mapping, and annotation of over 28,000 cDNA clones from japonica rice. Science. 2003;301:376–379. doi: 10.1126/science.1081288. [DOI] [PubMed] [Google Scholar]
- 6.Lu T, Huang X, Zhu C, Huang T, Zhao Q, Xie K, Xiong L, Zhang Q, Han B. RICD: a rice indica cDNA database resource for rice functional genomics. BMC Plant Biol. 2008;8:118. doi: 10.1186/1471-2229-8-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sakurai T, Kondou Y, Akiyama K, Kurotani A, Higuchi M, Ichikawa T, Kuroda H, Kusano M, Mori M, Saitou T, et al. RiceFOX: a database of Arabidopsis mutant lines overexpressing rice full-length cDNA that contains a wide range of trait information to facilitate analysis of gene function. Plant Cell Physiol. 2011;52:265–273. doi: 10.1093/pcp/pcq190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Krishnan A, Guiderdoni E, An G, Hsing YC, Han C, Lee MC, Yu S-M, Upadhyaya N, Ramachandran S, Zhang Q, et al. Mutant resources in rice for functional genomics of the grasses. Plant Physiol. 2009;149:165–170. doi: 10.1104/pp.108.128918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang H, Zhang D, Wang M, Sun J, Qi Y, Li J, Wei X, Han L, Qiu Z, Tang S, et al. A core collection and mini core collection of Oryza sativa L. in China. Theor. Appl. Genet. 2011;122:49–61. doi: 10.1007/s00122-010-1421-7. [DOI] [PubMed] [Google Scholar]
- 10.Li X, Yan W, Agrama H, Hu B, Jia L, Jia M, Jackson A, Moldenhauer K, McClung A, Wu D. Genotypic and phenotypic characterization of genetic differentiation and diversity in the USDA rice mini-core collection. Genetica. 2010;138:1221–1230. doi: 10.1007/s10709-010-9521-5. [DOI] [PubMed] [Google Scholar]
- 11.Gu H, Zhu P, Jiao Y, Meng Y, Chen M. PRIN: a predicted rice interactome network. BMC Bioinfo. 2011;12:161. doi: 10.1186/1471-2105-12-161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kurata N, Yamazaki Y. Oryzabase. An integrated biological and genome information database for rice. Bioinformatics. 2006;140:12–17. doi: 10.1104/pp.105.063008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pan B, Sheng J, Sun W, Zhao Y, Hao P, Li X. OrysPSSP: a comparative platform for small secreted proteins from rice and other plants. Nucleic Acids Res. 2013;41:D1192–D1198. doi: 10.1093/nar/gks1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sato Y, Namiki N, Takehisa H, Kamatsuki K, Minami H, Ikawa H, Ohyanagi H, Sugimoto K, Itoh J-I, Antonio BA, et al. RiceFREND: a platform for retrieving coexpressed gene networks in rice. Nucleic Acids Res. 2013;41:D1214–D1221. doi: 10.1093/nar/gks1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sato Y, Takehisa H, Kamatsuki K, Minami H, Namiki N, Ikawa H, Ohyanagi H, Sugimoto K, Antonio BA, Nagamura Y. RiceXPro version 3.0: expanding the informatics resource for rice transcriptome. Nucleic Acids Res. 2013;41:D1206–D1213. doi: 10.1093/nar/gks1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang D, Xia Y, Li X, Hou L, Yu J. The Rice Genome Knowledgebase (RGKbase): an annotation database for rice comparative genomics and evolutionary biology. Nucleic Acids Res. 2013;41:D1199–D1205. doi: 10.1093/nar/gks1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang Q, Li J, Xue Y, Han B, Deng XW. Rice 2020: a call for an international coordinated effort in rice functional genomics. Mol. Plant. 2008;1:715–719. doi: 10.1093/mp/ssn043. [DOI] [PubMed] [Google Scholar]
- 18.Salimi N, Vita R. The biocurator: connecting and enhancing scientific data. PLoS Comput. Biol. 2006;2:e125. doi: 10.1371/journal.pcbi.0020125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gaudet P, Arighi C, Bastian F, Bateman A, Blake JA, Cherry MJ, D’Eustachio P, Finn R, Giglio M, Hirschman L, et al. Recent advances in biocuration: meeting report from the Fifth International Biocuration Conference. Database. 2012;2012:bas036. doi: 10.1093/database/bas036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Van Auken K, Fey P, Berardini TZ, Dodson R, Cooper L, Li D, Chan J, Li Y, Basu S, Muller H-M, et al. Text mining in the biocuration workflow: applications for literature curation at WormBase, dictyBase and TAIR. Database. 2012;2012:bas040. doi: 10.1093/database/bas040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li D, Berardini TZ, Muller RJ, Huala E. Building an efficient curation workflow for the Arabidopsis literature corpus. Database. 2012;2012:bas047. doi: 10.1093/database/bas047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lu Z, Hirschman L. Biocuration workflows and text mining: overview of the BioCreative 2012 Workshop Track II. Database. 2012;2012:bas043. doi: 10.1093/database/bas043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lu Z, Hirschman L. Biocuration workflows and text mining: overview of the BioCreative 2012 Workshop Track II. Database. 2012;2012:bas043. doi: 10.1093/database/bas043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Arighi CN, Carterette B, Cohen KB, Krallinger M, Wilbur WJ, Fey P, Dodson R, Cooper L, Van Slyke CE, Dahdul W, et al. An overview of the BioCreative 2012 Workshop Track III: interactive text mining task. Database. 2013;2013:bas056. doi: 10.1093/database/bas056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Van Landeghem S, De Bodt S, Drebert ZJ, Inzé D, Van de Peer Y. The potential of text mining in data integration and network biology for plant research: a case study on Arabidopsis. Plant Cell. 2013;25:794–807. doi: 10.1105/tpc.112.108753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Plant Ontology and Consortium. The Plant Ontology Consortium and plant ontologies. Comp. Funct. Genomics. 2002;3:137–142. doi: 10.1002/cfg.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jaiswal P, Ware D, Ni J, Chang K, Zhao W, Schmidt S, Pan X, Clark K, Teytelman L, Cartinhour S, et al. Gramene: development and integration of trait and gene ontologies for rice. Comp. Funct. Genomics. 2002;3:132–136. doi: 10.1002/cfg.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Walls RL, Athreya B, Cooper L, Elser J, Gandolfo MA, Jaiswal P, Mungall CJ, Preece J, Rensing S, Smith B, et al. Ontologies as integrative tools for plant science. Am. J. Bot. 2012;99:1263–1275. doi: 10.3732/ajb.1200222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kawahara Y, de la Bastide M, Hamilton JP, Kanamori H, McCombie WR, Ouyang S, Schwartz DC, Tanaka T, Wu J, Zhou S, et al. Improvement of the Oryza sativa Nipponbare reference genome using next generation sequence and optical map data. Rice. 2013;6:1–10. doi: 10.1186/1939-8433-6-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lamesch P, Berardini TZ, Li D, Swarbreck D, Wilks C, Sasidharan R, Muller R, Dreher K, Alexander DL, Garcia-Hernandez M, et al. The Arabidopsis Information Resource (TAIR): improved gene annotation and new tools. Nucleic Acids Res. 2012;40:D1202–D1210. doi: 10.1093/nar/gkr1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cooper L, Walls RL, Elser J, Gandolfo MA, Stevenson DW, Smith B, Preece J, Athreya B, Mungall CJ, Rensing S, et al. The Plant Ontology as a tool for comparative plant anatomy and genomic analyses. Plant Cell Physiol. 2013;54:e1. doi: 10.1093/pcp/pcs163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Gene Ontology: tool for the unification of biology. Nat. Genet. 2011;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Punta M, Coggill PC, Eberhardt RY, Mistry J, Tate J, Boursnell C, Pang N, Forslund K, Ceric G, Clements J, et al. The Pfam protein families database. Nucleic Acids Res. 2012;40:D290–D301. doi: 10.1093/nar/gkr1065. [DOI] [PMC free article] [PubMed] [Google Scholar]




