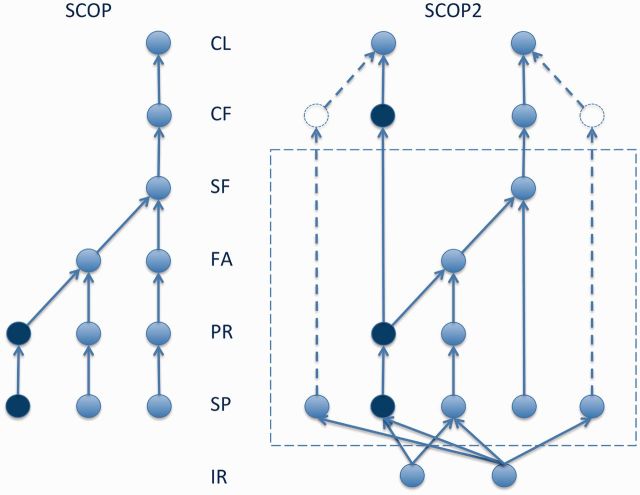
Figure 1.
SCOP and SCOP2 graphs compared. (Left) A section of the SCOP hierarchical tree, showing the classification into the six obligatory levels: Protein species (SP), Protein (PR), Family (FA), Superfamily (SF), Fold (CF) and Class (CL). The homologous proteins with distinct folds, e.g., the Cro repressors (25), are compulsorily assigned in the same family and so progressively to the same superfamily, fold and class. There is an obligatory node at every level from the root to a leaf, even if such node does not represent any actual relationship, e.g., ‘singleton’ protein family consisting of a single protein that only has a relationship with itself. (Right) In SCOP2, the structural and evolutionary relationships are separated, allowing the classification of the homologous proteins into different folds and structural classes while keeping them in the same evolutionary family and superfamily. Non-obligatory single-child nodes are skipped to emphasize that relatives of a ‘singleton’ protein exist only at the superfamily level. The new category specific to SCOP2, 'other' relationships (IR), is also shown on the graph. These relationships include but are not limited to non-hierarchical relationships between homologous and non-homologous proteins with different folds sharing a large common substructure or motif (see also Figure 2).