Figure 1.
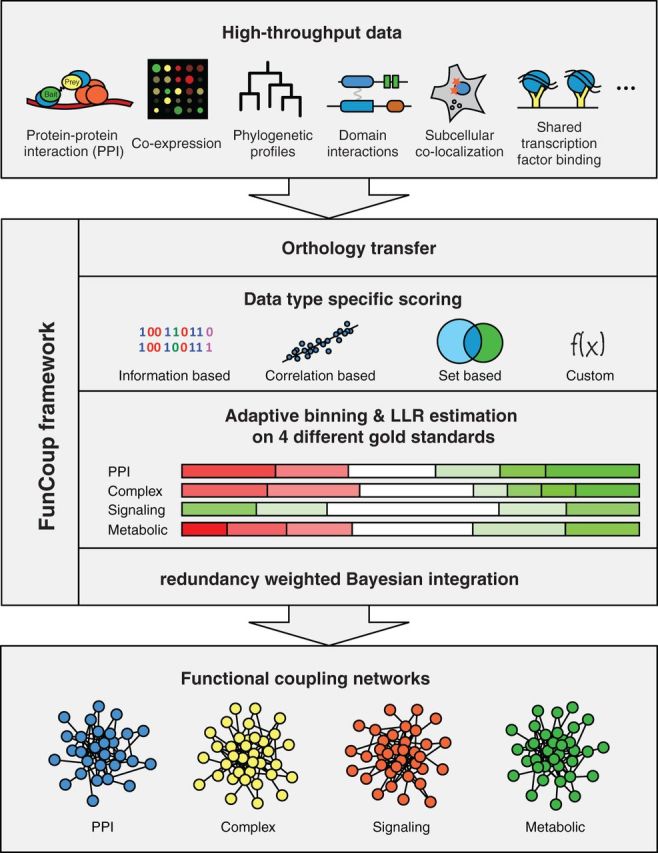
Overview of the FunCoup framework. FunCoup integrates nine different evidence types. For each evidence type, a unique scoring function is defined. The scoring functions are applied to the data of known associated gene pairs and random background pairs. The scores for the known and random pairs are used to partition the score range for each dataset by an adaptive binning procedure. The binning procedure identifies score ranges that are significantly enriched or depleted for known couplings. For each bin, the LLR between a known coupled pair and a background pair is estimated. In the next step, the scores for all possible gene pairs are calculated and translated into the corresponding LLRs. For each gene pair, the LLRs for each data type are combined by redundancy-weighted Bayesian integration. If the resulting FBS surpasses a threshold, a link is introduced for the gene pair. This way a genome-wide functional coupling network is created. FBSs are converted into more convenient confidence scores that range between 0 and 1 by using an alternative form of Bayes’ rule and assuming a prior interaction probability of 0.001. FunCoup has four different sets of known gene pairs representing different classes of functional couplings. The LLRs are estimated independently for each of these classes and four different networks are created. These are normally combined for searching purposes by keeping the maximum link strength, but they can also be used individually. FunCoup not only uses data from the species itself but also transfers data between species using orthology assignments from InParanoid. This is done before the LLRs are calculated; hence, data from a different species are evaluated the same way as data from the same species.
