Figure 3.
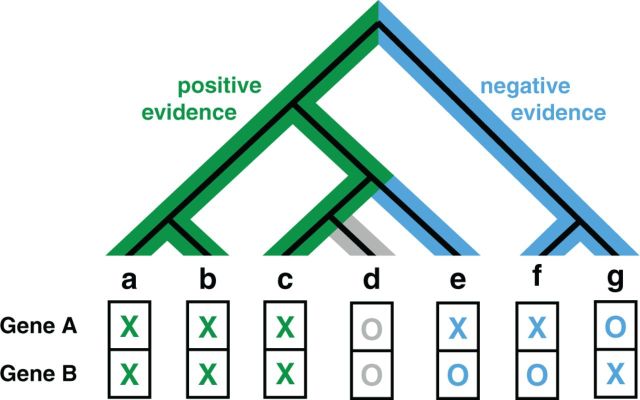
FunCoup 3.0 introduces a novel heuristic method for estimating PHP similarity. A neighbor-joining species tree is created for all except six eukaryotes in InParanoid using inter-species distances derived from the fraction of genes with orthologs as described in Berglund et al. (34). This tree is then rooted at the species for which the couplings are predicted. The PHP score has two components: the positive evidence and the negative evidence. The figure shows a simplified example with seven species. For the positive evidence, the score is the branch length of the green highlighted subtree where both genes have orthologs in InParanoid divided by the total branch length of the full tree. Whereas co-conservation provides positive evidence, negative evidence can be drawn from species where only one of the two genes has orthologs. The negative evidence score is therefore the sum of branch lengths of mutually exclusive species, highlighted in blue, divided by the total branch length of the tree. Branches that are covered by the positive evidence subtree are excluded from the negative evidence calculation, e.g. for species e, only the branch to the ancestor of c, d and e is included. This corresponds to the simplified assumption that genes can only be lost but not regained, and because species c has orthologs to gene A and gene B, we assume that the orthologs were also present at all ancestors of c. Both positive and negative scores have a value range between 0 and 1 and their sum is bound to maximum of 1. The two scores are binned into six fixed-size bins and an LLR is learned for every combination of a positive and a negative evidence bin. The score bins for positive and negative evidence span a 6 × 6 triangular matrix with 21 bins, which is significantly less than the 1024 bins for PHP in the previous release.
