Figure 1. CHD5 Chromatin Remodeling Protein.
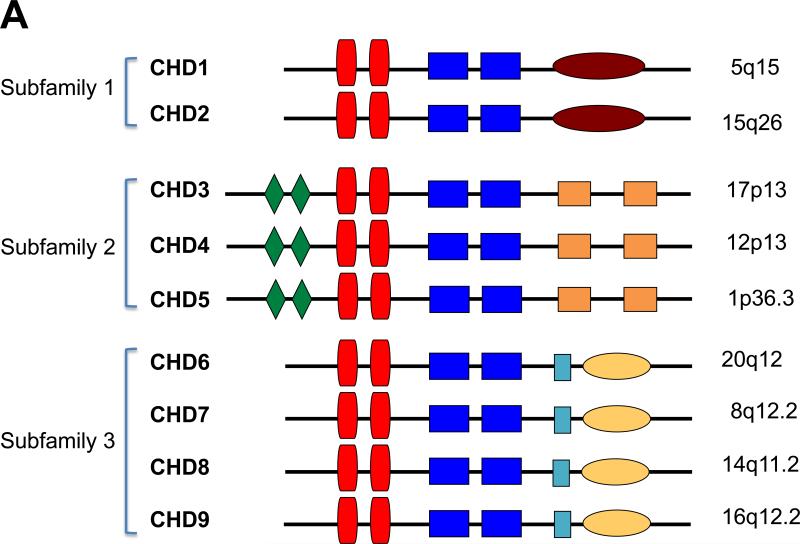
A. CHD family. Shown are diagrammatic representations of the nine members of the CHD (Chromodomain-Helicase-DNA binding) family of proteins. They are divided into three subfamilies based on sequence and structural homology as well as function. The conserved motifs are shown: green diamonds = PHD finger domains; red vertical boxes = chromodomains; blue rectangular boxes = split SNF2-like helicase/ATPase domain; burgundy oval = DNA binding motif; orange boxes = domains of unknown function (DUF1, DUF2); light blue box = SANT domain; yellow oval = BRK domains. The chromosomal locations of the nine CHD family members are shown to the right.
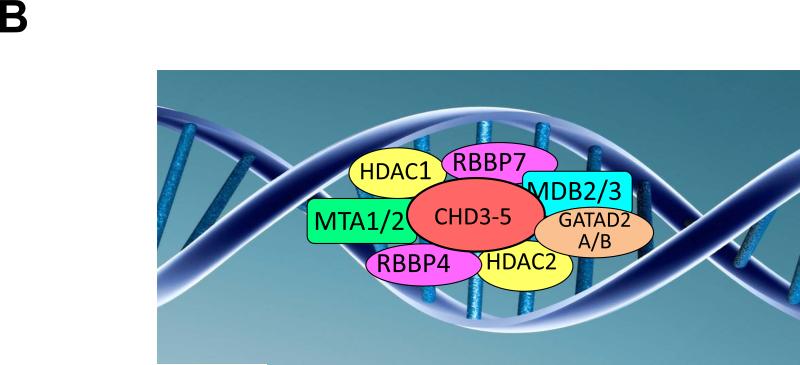
B. CHD5 NuRD complex. Shown is a diagrammatic representation of the hypothetical CHD5-NuRD complex, which is identical to a CHD4-NuRD complex. The canonical components of this complex are HDAC1 and HDAC2; MTA1 and MTA2; RBBP4 and RBBP7; GATA2DA and GATA2DB; and MDB2 and MDB3.
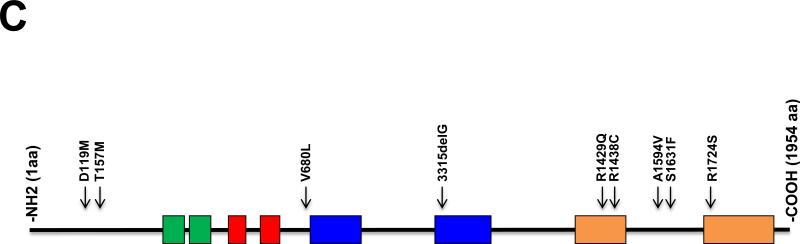
C. CHD5 mutations. Shown is a diagram of the CHD5 protein with conserved motifs, as indicated in 1A. The relative position of 9 CHD5 mutations in various cancers is listed in Table 1 and shown on this diagram. All of the mutations (8 of the 9) involved highly conserved amino acids (see Supplemental Figure 1). The G base pair deletion at nucleotide position 3315 changed the reading frame and led to a premature stop codon.