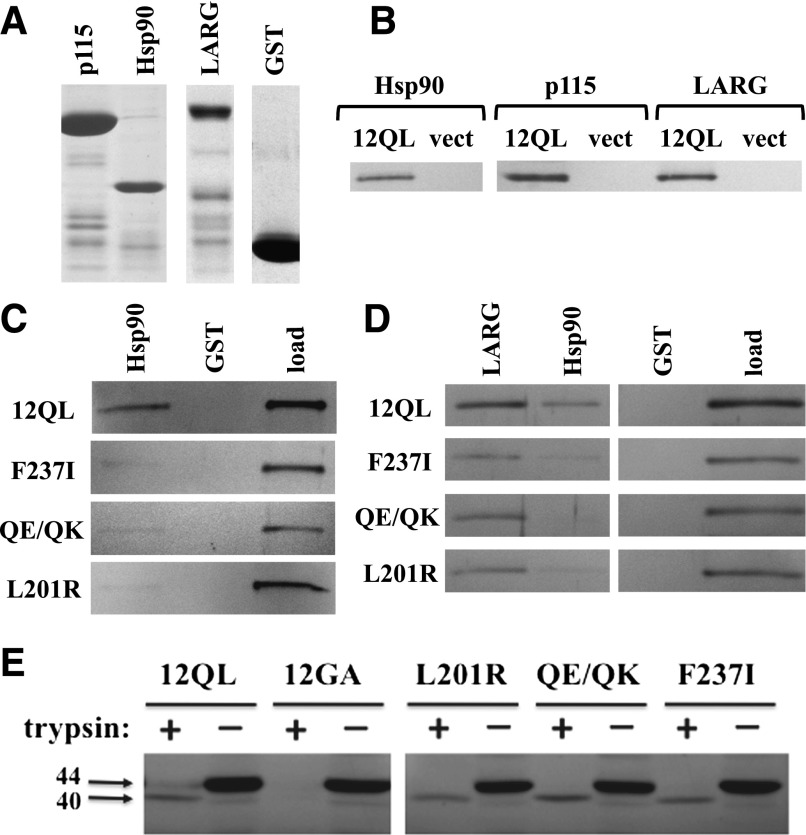
Fig. 3.
Effector binding and conformational activation of SRE-uncoupled class-distinctive Gα12 mutants. (A) GST fusions of the N-terminal 252 amino acids of p115RhoGEF (p115), the C-terminal 107 residues of cytoplasmic Hsp90, and the region spanning Leu320 to Arg606 of LARG were analyzed by SDS-PAGE and Coomassie blue staining. Unmodified GST was analyzed in parallel. (B) Coprecipitation experiments using these GST fusion proteins were performed on detergent extracts from HEK293 cells expressing myc-tagged, constitutively active Gα12 (12QL) or empty pcDNA3.1 (vect). (C) Coprecipitations of indicated Gα12 mutants by immobilized Hsp90 are shown. For each panel, the left and center lanes harbor samples in which Sepharose-bound GST-Hsp90 or GST (indicated at top) were used to precipitate fractions from HEK293 extracts harboring myc-Gα12QL (12QL) or the mutants indicated at left. The right lane of each panel (load) harbors 5% of the HEK293 extract set aside before coprecipitation, and band intensities at ∼44 kDa were quantified and used to normalize the coprecipitated bands for Gα12 variants indicated here, as well as others (Table 1). A representative of three independent experiments is shown. (D) Coprecipitation of myc-Gα12QL and its indicated mutants by multiple effectors are shown. The indicated GST fusions of LARG and Hsp90, plus GST with no adduct (GST) are indicated, along with the load for each sample. Data shown are a representative of three independent experiments. (E) Trypsin protection assays were performed on the indicated class-distinctive mutants, plus constitutively activated (12QL) and inactivated (12GA) myc-Gα12. HEK293 cells grown in 10-cm plates were transfected with 10 µg of plasmid DNA, and after 40 hours, cell lysates were subjected to tryptic proteolysis as described in Materials and Methods. Molecular weights are indicated at left in kilodaltons. Results shown are one representative of three independent experiments.