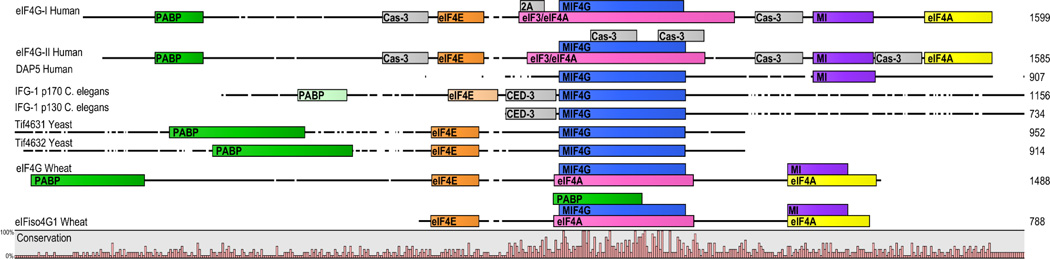
Figure 1. Comparisons of eIF4G across species.
Protein structures of eIF4G for different species aligned using the most highly conserved middle domain of eukaryotic initiation factor 4G (MIF4G; blue). Percent sequence conservation across the forms is indicated at the bottom of the alignment. DAP5 was included based on homology with both human eIF4G-I and –II. Conserved binding domains for eIF4E (orange), eIF3 and/or eIF4A (pink; yellow for second eIF4A binding site), and PABP (green) are shown. Putative PABP (light green) and eIF4E (light orange) binding sites are also indicated in the C. elegans IFG-1 isoform A and D (p170 and p130 respectively). The conservation of putative binding domains for eIF4A and eIF3, although presumably present, were not definitively identified in C. elegans and yeast. Cleavage sites for protease 2A, caspase-3, and caspase CED-3 (C. elegans) are shown in grey. The MA-3 and eIF4G domain (MI; purple) represents a homologous sequence between human and wheat eIF4G isoforms which has not been identified in C. elegans or yeast. See Supplemental Table 1 for amino acid binding sites. CLC Workbench 6.8.2 alignment.