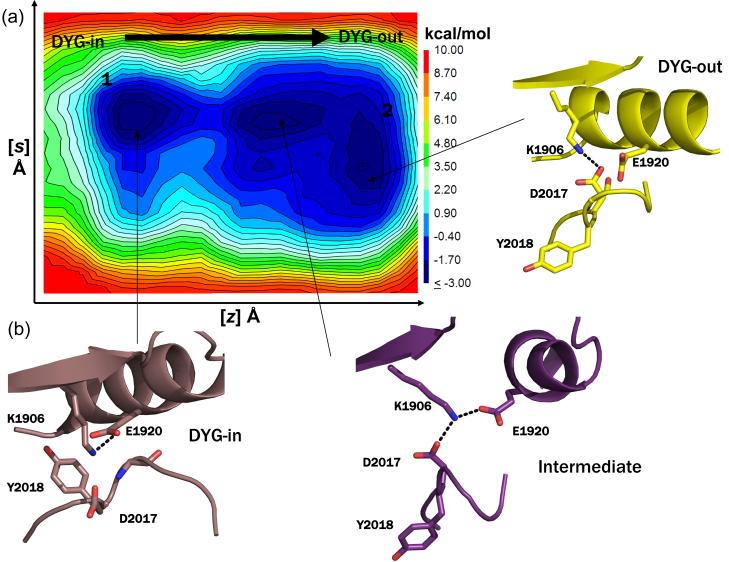
Figure 4.
(a) Representative free energy surface (FES) generated from 250 ns metadynamics simulation of WT LRRK2 generated by following the opening and closing of the enzyme involving the ATP binding β-sheet domain and C-helix as the first collective variable (z) and the motion of the center of mass of the activation loop as the second collective variable (s) as the kinase switches between active and inactive form. The FES shows that opening and closing motion of the enzyme occurs unhindered and the activation loop can switch between the active and inactive form easily (a contagious low energy path connects the two conformations forms). (b) Snapshots of structures extracted from local clustering of conformations within the simulation corresponding to the three local minima observed during the course of the simulation are shown here. The states 1 and 2 correspond to the active (DYG-in) and inactive (DYG-out) states of the enzyme. An intermediate metastable transition is observed, where K1906 makes a shared hydrogen bond with both E1920 (C-helix) and D2017 (DYG-motif).