Abstract
Chemoresistance is one of the major reasons for the failure of anticancer chemotherapy in treating advanced stage cancer. The mechanism of chemoresistance to fluoropyrimidines and antifolates has been extensively investigated in the past 40 years. It has been well established that thymidylate synthase (TYMS, TS) and dihydrofolate reductase (DHFR) are two major targets for fluoropyrimidines and antifolates, respectively. The regulatory mechanism of TS and DHFR expression is rather complex involving transcriptional, post-transcriptional and translational regulations. Our recent understanding of the chemoresistance mechanism has been extended beyond the simple one target/drug view. In this review, we will focus on the recent advancement of non-coding microRNAs (miRNAs) in contributing to the regulations of TS and DHFR expression, and to the chemoresistance mechanism of fluoropyrimidines and antifolates.
Keywords: Translational control, microRNA, thymidylate synthase, dihydrofolate reductase, 5-fluorouracil, methotrexate, chemoresistance
INTRODUCTION
Fluoropyrimidines (e.g. 5-fluorouracil, S-1) and antifolates (methotrexate, pemetrexed, raltitrexed) represent two major rationally designed anticancer drugs in the late 1950s and are still the cornerstones of anticancer chemotherapy after a half century [1–3]. Over the years, extensive efforts from many laboratories have contributed to the understanding of both molecular and cellular mechanism of anti-tumor effects of these two classes of drugs [4]. Because it is difficult to develop novel anti-cancer drugs, large amount of efforts have been contributed to enhance the efficacy of existing compounds via various strategies. These strategies were established based on the understanding of the mechanisms of drug action, target expression, and pathways. A number of adjuvant strategies have developed to further enhance the response and survival rate.
It is well established that 5-fluorouracil (5-FU) targets a critical enzyme TS [5]. TS is a folate-dependent enzyme that catalyzes the reductive methylation of dUMP by 5,10-methylenetetrahydrofolate to form dTMP and dihydrofolate. Because the TS-catalyzed enzymatic reaction provides the sole intracellular de novo source of thymidylate, an essential precursor for DNA biosynthesis, this enzyme has been an important target for cancer chemotherapy for over a half century. 5-FU also directly incorporates into DNA and RNA to trigger cell death [6, 7].
The key target for antifolate based therapy is DHFR [8]. DHFR catalyzes the NADPH-dependent reduction of dihydrofolate to tetrahydrofolate, a key intermediate molecule in one-carbon transfer reactions. DHFR plays a key role in folate homeostasis, and provides the one-carbon carrier units that are required for the de novo synthesis of purines, thymidylate, and certain amino acids. As a result, targeting DHFR represents another important therapeutic strategy in antifolate-based cancer chemotherapy [9].
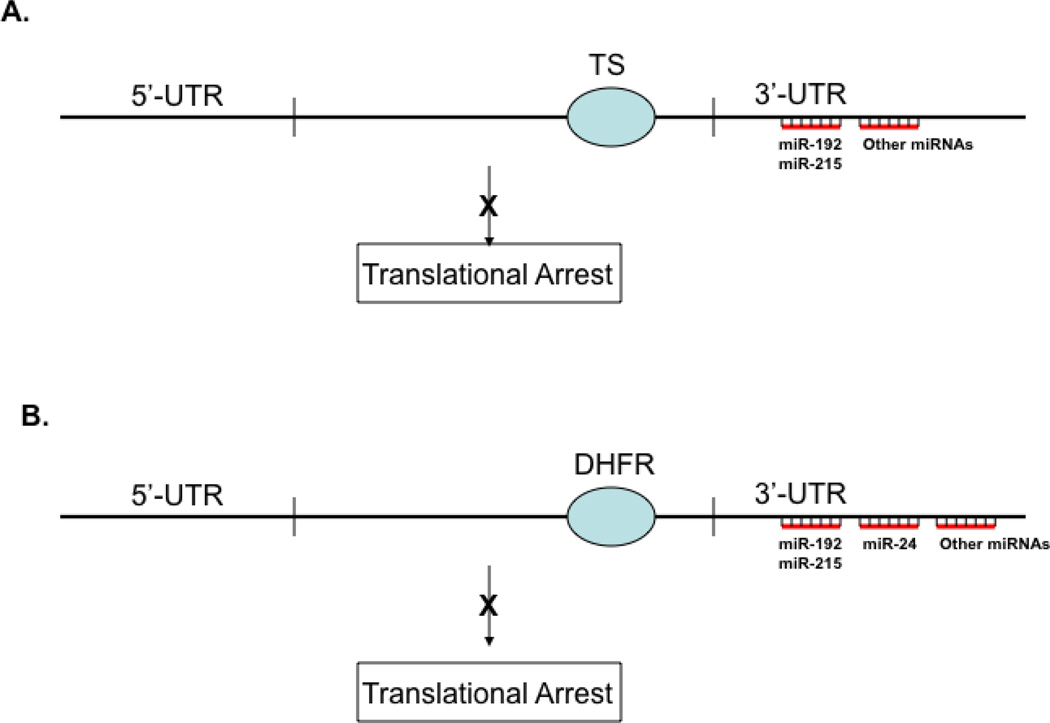
The molecular mechanisms of regulation of TS and DHFR expression are rather complex. TS and DHFR were both transcriptionally and post-transcriptionally regulated. With regards to translational control, both TS and DHFR act as RNA binding proteins to autoregulate their own expression [10–12]. It has also been reported that the polymorphisms at both 5’- and 3’-UTR regions of TS mRNA also influence TS protein expression [13–15]. This review will focus on the new recent findings that non-coding microRNAs (miRNAs) are also involved in regulating TS (Fig. 1A) and DHFR (Fig. 1B) expression and such regulation contribute to the chemoresistance mechanism to TS and DHFR inhibitors.
Fig. (1).
Translational regulation of TS expression. TS expression is auto-regulated by its own protein by direct binding to its own mRNA at the coding region. The translation of TS protein was also directly regulated by miRNAs (e.g. miR-192, miR-215) by interacting with the 3’-UTR regions of TS mRNA (1A). Translational regulation of DHFR expression. DHFR expression is auto-regulated by its own protein by direct binding to its own mRNA at the coding region. The translation of DHFR protein was also directly regulated by miRNAs (e.g. miR-192, miR-215, miR-24) by interacting with the 3’-UTR regions of DHFR mRNA (1B).
miRNA BIOGENESIS
miRNAs are a class of non-coding RNAs (e.g. siRNA, piRNA) with crucial regulatory functions, as demonstrated by the recent Nobel Prize recognition of early breakthrough of RNA interference in this field [16]. Ambros and Ruvkun’s group made a landmark discovery in 1993 that a small non-coding RNA, lin-4, impacted the development of Caenorhabditis elegans [17, 18]. The impact of miRNA in cancer was discovered a decade later by demonstrating a direct link of downregulation of miR-15 and miR-16 in chronic lymphocytic leukemia [19]. The impact of miRNAs in cancer has now become a new frontier in cancer research. Currently, more than 1,000 mammalian miRNAs have been identified by cloning and sequencing approaches [20], including hundreds in humans [21].
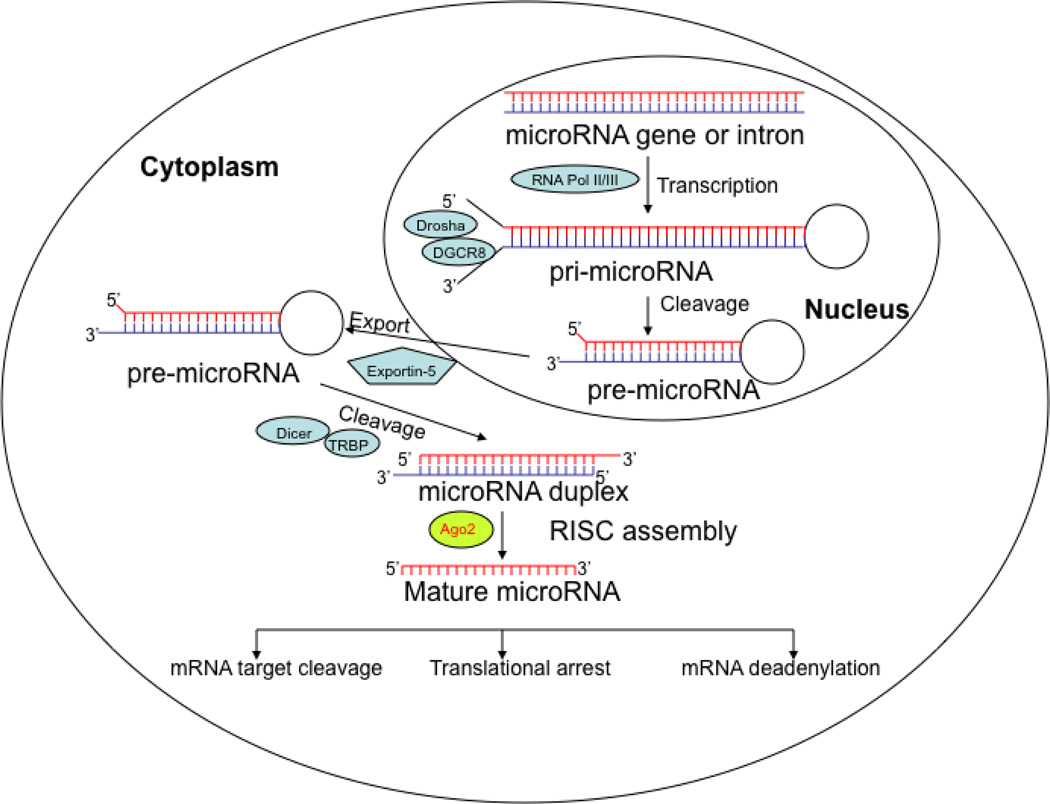
miRNAs are a class of small non-coding RNA molecules of 20–22 nucleotides in length, which are processed from larger pre-miRNAs by the RNase III enzyme Dicer (DICER1) into miRNA duplexes [22] (Fig. 2). One strand of this duplex associates with the RNA-induced silencing complex (RISC), whereas the other strand is generally degraded by cellular nucleases [22]. The miRNA–RISC complex binds to specific messenger RNA (mRNA) targets, leading to translational repression or cleavage of these mRNAs. Thus, miRNAs modulate protein expression by promoting RNA degradation, inhibiting mRNA translation, and in some cases, affecting transcription. Although miRNA-mediated mRNA degradation occurs in mammals, most mammalian miRNAs are thought to repress target gene expression at the translational level [17, 23, 24] via imperfect base-pairing to the 3’-untranslated regions (3’-UTRs) of their target mRNAs. This form of translational regulation provides the cell with a more precise, immediate and energy-efficient way of controlling the expression of a given protein [25] as it induces rapid changes in protein synthesis without excess transcriptional activation and subsequent steps in mRNA processing. Additionally, translational control of gene expression has the advantage of being readily reversible, providing the cell with great flexibility in responding to various cytotoxic stresses.
Fig. (2).
Primary miRNA (pri-miRNA) is transcribed by RNA polymerase II or III and subsequently cleaved to precursor miRNA (pre-miRNA) by Drosha-DGCR8 (Pasha) complex in the nucleus. The pre-miRNA is exported to the cytoplasm by Exportin protein complex. The pre-miRNA is further cleaved by RNase Dicer complex with RNA binding protein TRBP to form mature length. The functional strand of the mature miRNA is loaded with Argonaute (Ago2) into the RNA-induced silencing complex (RISC) to prevent protein synthesis through mRNA cleavage, translational arrest or deadenylation.
To fully understand gene expression changes in cancer, it is essential to know not only the levels of individual mRNAs, but also the extent to which mRNAs are translated into their corresponding proteins and the miRNAs that regulate these processes. Thus, post-transcriptional and translational controls mediated by miRNAs have come under increasing scrutiny in cancer research. Mounting evidence suggests that miRNAs can function as either oncogenes or tumor suppressor genes. As a result, some of them will have potential as therapeutic targets.
5-Fluorouracil (5-FU), miRNA, Chemoresistance, and Colon Cancer Stem Cells
As mentioned in the beginning, 5-FU-based chemotherapy is one of the main treatment options for advanced metastatic colorectal cancer and other solid tumors for well over half a century. However, despite the steady improvement in response rate with various modulation strategies such as leucovorin (LV), methotrexate (MTX), and Oxaliplatin, many patients still go through 5-FU-based chemotherapy without any benefit. A number of key factors have attributed to the lack of response and survival benefit. One of the major causes is largely due to the heterogeneity of the tumor cells, in particular, slow growing tumor stem like cells that are resistant to anticancer drugs [26–28]. Despite the extensive investigations to discover predictive response biomarkers for 5-FU-based treatment, the predictive power of TS for 5-FU-based chemotherapy is still currently under debate [29, 30]. Further, studies evaluating TS, thymidine phosphorylase (TP), and dihydropyrimidine dehydrogenase (DPD), a catabolic enzyme of 5-FU as predictive biomarkers, were unable to confirm the high response rates reported by several retrospective studies [31]. It is clear that the search continues for biomarkers that can be used, either alone or in combination with existing biomarkers such as TS and DPD, to predict the likely response to 5-FU-based treatment. Chemoresistance is one of the major factors for the failure of chemotherapy. One effect behind chemoresistance is that chemotherapeutic agents, which primarily affect rapid-proliferating cells, are mostly ineffective against the more slowly-proliferating and/or quiescent cancer stem cells, allowing a tumor to reconstitute itself once therapy has ceased. The broad influence and reversible nature of miRNA on gene expression have provided us, as well as other researchers, with experimental evidence to support that miRNAs may offer new insights to this resistance mechanism, in particular with 5-FU-based colorectal cancer chemotherapy. Ultimately, the information we gained may help to develop new strategies to overcome chemoresistance and to better predict clinical outcomes.
miRNA, p53 and Colorectal Cancer
p53 is one of the most frequently mutated and/or deleted tumor suppressor genes in colorectal cancer and other tumor types: nearly half of all colorectal cancer patients carry p53 mutations or deletions [32]. Until recently, it has been thought that the effects of p53 loss were primarily related to its function in transcriptional regulation, but it is now appreciated that post-transcriptional and translational controls are equally important. Our own research efforts began by systematically investigating which miRNAs are influenced by the p53 tumor suppressor and, by extension, p53 loss. In 2006 we first reported a regulatory relationship between p53 and a number of miRNAs [33], and discovered that nearly half of the 328 miRNA putative promoter regions, including the promoters for miR-34s, miR-192, miR-215, and miR-26a, contain one or more p53 binding sites [33]. Subsequently, several groups have demonstrated that miR-34a, which acts as a tumor-suppressor by targeting several cell-cycle genes, is regulated directly by p53 [34–36]. In addition, we and two other groups provided further direct evidence that miR-192 is another miRNA that is both regulated by p53 and capable of inducing cell-cycle arrest [37–39].
Our recent efforts have focused on miRNAs that suppress the expression of TS and DHFR, two important chemotherapeutic targets. We have discovered that two miRNAs we have previously identified as being regulated by p53, miR-192 and miR-215, target both TS and DHFR mRNA at the 3’-UTRs to suppress protein translation [37, 40] (Fig. 1). Additionally, Mishra et al. reported that miR-24 also regulates DHFR expression [41], and demonstrated that 829C→T, a naturally occurring SNP near the miR-24 binding site in the 3' UTR of DHFR, interferes with miR-24 function and, resulting in DHFR overexpression and methotrexate resistance in colorectal tumors [42]. Further studies revealed that miR-24 is a potential tumor suppressor capable of reducing tumor cell proliferation in a p53-independent fashion and mediating several key cell cycle-related genes such as p21, E2F, Myc and other cell cycle control genes [43, 44]. It is also tempting to reason that nature builds such redundancy to adopt various growth stress conditions by utilizing several different miRNAs to quickly modulate protein expression.
The involvement of several miRNAs in p53 mediated pathways provides a unique advantage for cells to adopt different acute changes in growth conditions. It is also quite possible that different miRNAs react to each unique growth condition change to ensure the precision of protein expression. The notion of different miRNAs (miR-24, miR-192, miR-215) capable of interacting with a particular mRNA transcript to modulate translation draws a conclusion that fine tuning translational efficiency of a particular mRNA is crucial for cells to quickly adapt to growth environmental changes with one or several different miRNAs.
The positive feedback mechanism between p53 and some of these miRNAs is clearly evidenced to be an important part of the regulatory function and networks of p53 mediated through miRNAs. Due to the broad impact of miRNA on regulating the translation rate, miRNAs may be responsible for the fine tuning of the tumor suppressor function of p53 under acute growth environmental changes including genotoxic stress. Therefore, we have reason to believe that modulation of miRNAs will have a broad impact in colorectal and other cancer types.
Impact of miRNAs in Cancer Stem Cells
Resistance to chemotherapeutic agents is a major reason for the failure of fluoropyrimidine-based anti-cancer treatment (e.g. FOLFOX). Many factors contribute to the resistance phenotype, especially in quiescent and slowly proliferating cancer stem cells [45–47]. Cancer stem cells are defined as the unique subpopulation of tumor cells that possess the ability to initiate tumor growth and sustain self-renewal as well as metastatic potential. Colon cancer stem cells are thought to be slowly dividing, relatively apoptosis-resistant and gradually differentiate into more highly proliferative colon cancer cells. Tumor stem cells are highly resistant to chemotherapeutic treatment due to the slow proliferating phenotype [28]. We have recently discovered that colon cancer stem cells maintain such phenotype with reduced TS and DHFR expressions regulated by elevated expression of miR-215 [48].
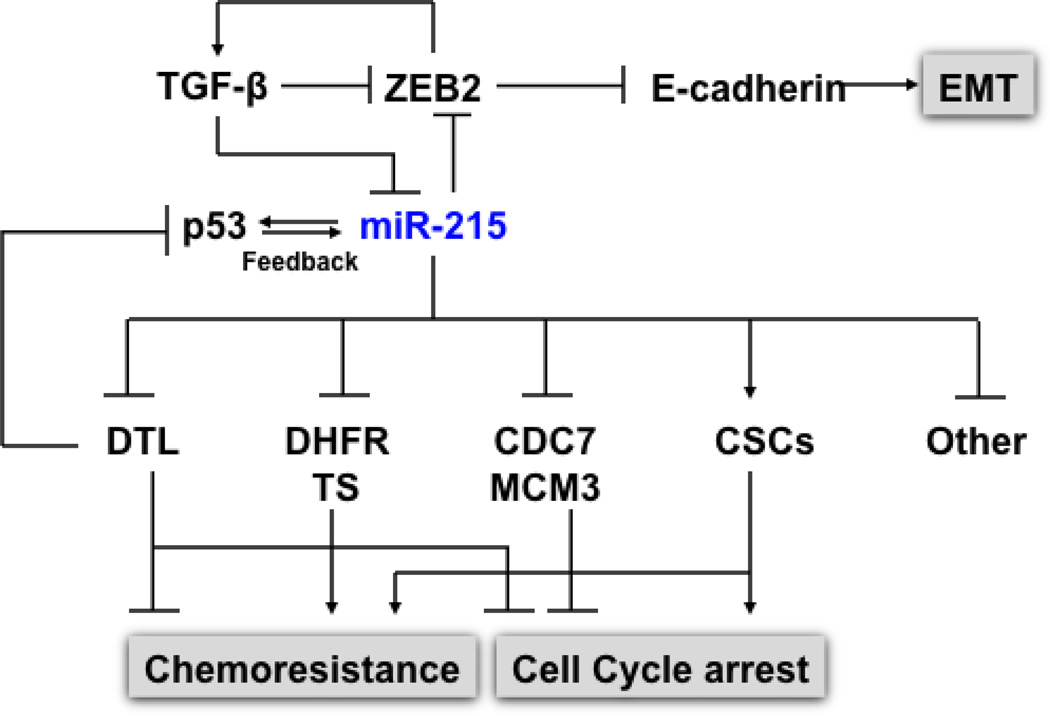
As mentioned briefly above, recent studies from our group have found that miR-215 directly targets the expression of both TS and DHFR [48]. Strikingly, our studies found that despite the reduction of TS and DHFR levels with elevated expression of miR-215, colon cancer cells became more resistant to the TS inhibitor Tomudex (TDX) and DHFR inhibitor methotrexate (MTX). Further investigation found that this effect was mainly due to the enhanced G2/M cell cycle check point control and reduced cell proliferation. The enhanced G2 check point control was mediated by the suppression of denticleless (DTL), one of the components of CUL4A-DDB1 E3 ubiquitin ligase complex [49], and essential for the early G2/M checkpoint. We discovered that the induction of p53 and p21 was a result of DTL suppression by miR-215, and that siRNA knockdown of DTL conferred chemoresistance to TDX and also triggered G2/M arrest in colon cancer cells. Taken together, this suggests the presence of a positive feedback loop between miR-215 and p53, mediated through the suppression of DTL (Fig. 3).
Fig. (3).
Impact of miR-215 on cell proliferation, cell cycle, chemoresistance and EMT. p53 and miR-215 forms a positive feedback regulatory mechanism to regulate cell cycle control. The expression of p53 was induced by the suppression of DTL, a critical component of the CUL4A-DDB1 E3 ubiquitin ligase complex, along with the p53 down stream gene p21. The elevated p53 in turn up-regulates the expression of miR-215. In addition to DTL, miR-215 also suppresses the expression of TS, DHFR, CDC7, MCM3, and MCM10. miR-215 triggers cell cycle arrest at the G2/M phase without triggering cell death, which contributes to chemoresistance to TS and DHFR inhibitors in CSCs. miR-215 also mediates EMT transition by targeting ZEB2 via TGFβ signaling pathway.
Epigenetic alterations contribute greatly to the dynamic and reversible phenotype of tumor cells and allow tumor cells to quickly adapt to the changes in tumor microenvironment. One key protein-regulating epigenetic alteration is histone deacetylase (HDAC). We have recently identified one of the key targets of miR-140 to be HDAC4 [50]. Ectopic expression of miR-140 reduced colon cancer cell proliferation and triggered cell cycle arrest. miR-140 over-expression also rendered colon cancer cells more resistant to TDX and MTX treatment. Further studies from our laboratory reveal that colon cancer stem cell populations, known to be highly resistant to 5-FU treatment, contain high levels of miR-215 and miR-140 [48, 50]. By blocking the activity of miR-140 using anti-miR-140 LNA oligonucleotides, we were able to sensitize colon cancer stem cells to 5-FU treatment [50].
These findings raise an interesting point: Previously, miRNAs have been classified as either pro- or antioncogenic. However, given the breadth of targets and pathways impacted by miRNA, it can target both oncogenes and tumor suppressor genes resulting in a net phenotypic outcome that is rather complex. For example, miR-215 and miR-140 appear at first glance to act as tumor suppressors because they can slow tumor growth and reduce tumor size in vivo. However, despite their anti-proliferative effect, both miRNAs produced cells resistant to chemotherapy. This may be a reason why we are able to cure mice by shrinking human tumor xenografts with various approaches. Most of these approaches and positive impacts, however, will not translate to human subjects as we were unable to monitor tumor relapse from the small population of chemoresistant tumor cells due to the short life span of the mice. Innovative strategies have to be developed to overcome such difficulties.
FUTURE PERSPECTIVES
Post-transcriptional control, miRNAs in particular, are now being recognized as important cellular processes, the alteration of which can contribute to a number of human diseases. miRNA-mediated regulation allows cells to fine-tune the expression of multiple genes and pathways via an energy-efficient mechanism that can be both quickly effected and readily reversed, providing an incredible degree of flexibility in the face of constantly changing conditions. This highly-adaptive system, however, is also inherited by these cells that become cancerous, contributing to chemoresistance and other survival mechanisms. In the present review, we summarized the role of miRNAs in colorectal cancer and chemoresistance mechanism. The incredible complexity of the miRNAs in gene regulation will certainly provide a host of future discoveries of new regulatory mechanisms ranging from the level of the individual genes to the vast regulatory networks. With the continual advancement of knowledge in the domains of miRNA, mRNA, protein expression, and computational biology, we come ever closer to fully understanding the complex miRNA-mediated regulatory pathways and networks in both cancer and normal cells. This knowledge, in turn, will undoubtedly continue to offer potential new therapeutic strategies and diagnostic/prognostic biomarkers for the treatment of colorectal cancer and other human diseases.
ACKNOWLEDGEMENTS
We appreciate the critical review by Ms. Sonya R. Lorrain. We apologize to our colleagues whose research was not cited in this review due to the focus, space limitations and timing. We like to acknowledge the support in part by Stony Brook University Translational Research Laboratory Start-up Fund (J. Ju), R01CA155019 (J. Ju) and R33CA147966 (J. Ju).
Footnotes
CONFLICT OF INTEREST
None.
REFERENCES
- 1.Chaudhuri NK, Montag BJ, Heidelberger C. Studies on fluorinated pyrimidines. III. The metabolism of 5-fluorouracil-2-C14 and 5-fluoroorotic-2-C14 acid in vivo. Cancer Res. 1958;18:318–328. [PubMed] [Google Scholar]
- 2.Holland JF. Methotrexate therapy of metastatic choriocarcinoma. Am. J. Obstet. Gynecol. 1958;75:195–199. doi: 10.1016/0002-9378(58)90571-4. [DOI] [PubMed] [Google Scholar]
- 3.Heidelberger C, Chaudhuri NK, Danneberg P, Mooren D, Griesbach L, Duschinsky R, Schnitzer RJ, Pleven E, Scheiner J. Fluorinated pyrimidines, a new class of tumour-inhibitory compounds. Nature. 1957;179:663–666. doi: 10.1038/179663a0. [DOI] [PubMed] [Google Scholar]
- 4.Sobrero AF, Aschele C, Bertino JR. Fluorouracil in colorectal cancer--a tale of two drugs: implications for biochemical modulation. J. Clin. Oncol. 1997;15:368–381. doi: 10.1200/JCO.1997.15.1.368. [DOI] [PubMed] [Google Scholar]
- 5.Danenberg PV. Thymidylate synthetase - a target enzyme in cancer chemotherapy. Biochim. Biophys. Acta. 1977;473:73–92. doi: 10.1016/0304-419x(77)90001-4. [DOI] [PubMed] [Google Scholar]
- 6.Bunz F, Hwang PM, Torrance C, Waldman T, Zhang Y, Dillehay L, Williams J, Lengauer C, Kinzler KW, Vogelstein B. Disruption of p53 in human cancer cells alters the responses to therapeutic agents. J. Clin. Invest. 1999;104:263–269. doi: 10.1172/JCI6863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schmittgen TD, Danenberg KD, Horikoshi T, Lenz HJ, Danenberg PV. Effect of 5-fluoro- and 5-bromouracil substitution on the translation of human thymidylate synthase mRNA. J. Biol. Chem. 1994;269:16269–16275. [PubMed] [Google Scholar]
- 8.Hillcoat BL, Swett V, Bertino JR. Increase of dihydrofolate reductase activity in cultured mammalian cells after exposure to methotrexate. Proc. Natl. Acad. Sci. USA. 1967;58:1632–1637. doi: 10.1073/pnas.58.4.1632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tai N, Schmitz JC, Liu J, Lin X, Bailly M, Chen TM, Chu E. Translational autoregulation of thymidylate synthase and dihydrofolate reductase. Front. Biosci. 2004;9:2521–2526. doi: 10.2741/1413. [DOI] [PubMed] [Google Scholar]
- 10.Ercikan E, Banerjee D, Waltham M, Schnieders B, Scotto KW, Bertino JR. Translational regulation of the synthesis of dihydrofolate reductase. Adv. Exp. Med. Biol. 1993;338:537–540. doi: 10.1007/978-1-4615-2960-6_109. [DOI] [PubMed] [Google Scholar]
- 11.Chu E, Takimoto CH, Voeller D, Grem JL, Allegra CJ. Specific binding of human dihydrofolate reductase protein to dihydrofolate reductase messenger RNA in vitro. Biochemistry. 1993;32:4756–4760. doi: 10.1021/bi00069a009. [DOI] [PubMed] [Google Scholar]
- 12.Chu E, Koeller DM, Casey JL, Drake JC, Chabner BA, Elwood PC, Zinn S, Allegra CJ. Autoregulation of human thymidylate synthase messenger RNA translation by thymidylate synthase. Proc. Natl. Acad. Sci. USA. 1991;88:8977–8981. doi: 10.1073/pnas.88.20.8977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Horie N, Takeishi K. Functional structure of the promoter region of the human thymidylate synthase gene and nuclear factors that regulate the expression of the gene. Nucleic Acids Symp. Ser. 1995;34:77–78. [PubMed] [Google Scholar]
- 14.Ulrich CM, Bigler J, Velicer CM, Greene EA, Farin FM, Potter JD. Searching expressed sequence tag databases: discovery and confirmation of a common polymorphism in the thymidylate synthase gene. Cancer Epidemiol. Biomarkers Prev. 2000;9:1381–1385. [PubMed] [Google Scholar]
- 15.Mandola MV, Stoehlmacher J, Zhang W, Groshen S, Yu MC, Iqbal S, Lenz HJ, Ladner RD. A 6 bp polymorphism in the thymidylate synthase gene causes message instability and is associated with decreased intratumoral TS mRNA levels. Pharmacogenetics. 2004;14:319–327. doi: 10.1097/00008571-200405000-00007. [DOI] [PubMed] [Google Scholar]
- 16.Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 17.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 18.Wightman B, Ha I, Ruvkun G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell. 1993;75:855–862. doi: 10.1016/0092-8674(93)90530-4. [DOI] [PubMed] [Google Scholar]
- 19.Calin GA, Dumitru CD, Shimizu M, Bichi R, Zupo S, Noch E, Aldler H, Rattan S, Keating M, Rai K, Rassenti L, Kipps T, Negrini M, Bullrich F, Croce CM. Frequent deletions and down-regulation of micro- RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc. Natl. Acad. Sci. USA. 2002;99:15524–15529. doi: 10.1073/pnas.242606799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 21.Cummins JM, He Y, Leary RJ, Pagliarini R, Diaz LA, Jr, Sjoblom T, Barad O, Bentwich Z, Szafranska AE, Labourier E, Raymond CK, Roberts BS, Juhl H, Kinzler KW, Vogelstein B, Velculescu VE. The colorectal microRNAome. Proc. Natl. Acad. Sci. USA. 2006;103:3687–3692. doi: 10.1073/pnas.0511155103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Esquela-Kerscher A, Slack FJ. Oncomirs - microRNAs with a role in cancer. Nat. Rev. Cancer. 2006;6:259–269. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- 23.Pillai RS, Bhattacharyya SN, Artus CG, Zoller T, Cougot N, Basyuk E, Bertrand E, Filipowicz W. Inhibition of translational initiation by Let-7 MicroRNA in human cells. Science. 2005;309:1573–1576. doi: 10.1126/science.1115079. [DOI] [PubMed] [Google Scholar]
- 24.Ruvkun G. Clarifications on miRNA and cancer. Science. 2006;311:36–37. doi: 10.1126/science.311.5757.36d. [DOI] [PubMed] [Google Scholar]
- 25.Dony C, Kessel M, Gruss P. Post-transcriptional control of myc and p53 expression during differentiation of the embryonal carcinoma cell line F9. Nature. 1985;317:636–639. doi: 10.1038/317636a0. [DOI] [PubMed] [Google Scholar]
- 26.Ricci-Vitiani L, Lombardi DG, Pilozzi E, Biffoni M, Todaro M, Peschle C, De Maria R. Identification and expansion of human colon-cancer-initiating cells. Nature. 2007;445:111–115. doi: 10.1038/nature05384. [DOI] [PubMed] [Google Scholar]
- 27.O'Brien CA, Pollett A, Gallinger S, Dick JE. A human colon cancer cell capable of initiating tumour growth in immunodeficient mice. Nature. 2007;445:106–110. doi: 10.1038/nature05372. [DOI] [PubMed] [Google Scholar]
- 28.Botchkina IL, Rowehl RA, Rivadeneira DE, Karpeh MS, Jr, Crawford H, Dufour A, Ju J, Wang Y, Leyfman Y, Botchkina GI. Phenotypic subpopulations of metastatic colon cancer stem cells: genomic analysis. Cancer Genomics Proteomics. 2009;6:19–29. [PubMed] [Google Scholar]
- 29.Salonga D, Danenberg KD, Johnson M, Metzger R, Groshen S, Tsao-Wei DD, Lenz HJ, Leichman CG, Leichman L, Diasio RB, Danenberg PV. Colorectal tumors responding to 5-fluorouracil have low gene expression levels of dihydropyrimidine dehydrogenase, thymidylate synthase, and thymidine phosphorylase. Clin. Cancer Res. 2000;6:1322–1327. [PubMed] [Google Scholar]
- 30.Showalter SL, Showalter TN, Witkiewicz A, Havens R, Kennedy EP, Hucl T, Kern SE, Yeo CJ, Brody JR. Evaluating the drug-target relationship between thymidylate synthase expression and tumor response to 5-fluorouracil. Is it time to move forward? Cancer Biol. Ther. 2008;7:986–994. doi: 10.4161/cbt.7.7.6181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Smorenburg CH, Peters GJ, van Groeningen CJ, Noordhuis P, Smid K, van Riel AM, Dercksen W, Pinedo HM, Giaccone G. Phase II study of tailored chemotherapy for advanced colorectal cancer with either 5-fluouracil and leucovorin or oxaliplatin and irinotecan based on the expression of thymidylate synthase and dihydropyrimidine dehydrogenase. Ann. Oncol. 2006;17:35–42. doi: 10.1093/annonc/mdj046. [DOI] [PubMed] [Google Scholar]
- 32.Hollstein M, Sidransky D, Vogelstein B, Harris CC. p53 mutations in human cancers. Science. 1991;253:49–53. doi: 10.1126/science.1905840. [DOI] [PubMed] [Google Scholar]
- 33.Xi Y, Shalgi R, Fodstad O, Pilpel Y, Ju J. Differentially regulated micro-RNAs and actively translated messenger RNA transcripts by tumor suppressor p53 in colon cancer. Clin. Cancer Res. 2006;12(Pt 1):2014–2024. doi: 10.1158/1078-0432.CCR-05-1853. [DOI] [PubMed] [Google Scholar]
- 34.He L, He X, Lim LP, de Stanchina E, Xuan Z, Liang Y, Xue W, Zender L, Magnus J, Ridzon D, Jackson AL, Linsley PS, Chen C, Lowe SW, Cleary MA, Hannon GJ. A microRNA component of the p53 tumour suppressor network. Nature. 2007;447:1130–1134. doi: 10.1038/nature05939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chang TC, Wentzel EA, Kent OA, Ramachandran K, Mullendore M, Lee KH, Feldmann G, Yamakuchi M, Ferlito M, Lowenstein CJ, Arking DE, Beer MA, Maitra A, Mendell JT. Transactivation of miR-34a by p53 broadly influences gene expression and promotes apoptosis. Mol. Cell. 2007;26:745–752. doi: 10.1016/j.molcel.2007.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Raver-Shapira N, Marciano E, Meiri E, Spector Y, Rosenfeld N, Moskovits N, Bentwich Z, Oren M. Transcriptional activation of miR-34a contributes to p53-mediated apoptosis. Mol. Cell. 2007;26:731–743. doi: 10.1016/j.molcel.2007.05.017. [DOI] [PubMed] [Google Scholar]
- 37.Song B, Wang Y, Kudo K, Gavin EJ, Xi Y, Ju J. miR-192 Regulates dihydrofolate reductase and cellular proliferation through the p53-microRNA circuit. Clin. Cancer Res. 2008;14:8080–8086. doi: 10.1158/1078-0432.CCR-08-1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Georges SA, Biery MC, Kim SY, Schelter JM, Guo J, Chang AN, Jackson AL, Carleton MO, Linsley PS, Cleary MA, Chau BN. Coordinated regulation of cell cycle transcripts by p53-inducible microRNAs, miR-192 and miR-215. Cancer Res. 2008;68:10105–10112. doi: 10.1158/0008-5472.CAN-08-1846. [DOI] [PubMed] [Google Scholar]
- 39.Braun CJ, Zhang X, Savelyeva I, Wolff S, Moll UM, Schepeler T, Ørntoft TF, Andersen CL, Dobbelstein M. p53-Responsive MicroRNAs 192 and 215 Are Capable of Inducing Cell Cycle Arrest. Cancer Res. 2008;68:10094–10104. doi: 10.1158/0008-5472.CAN-08-1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Song B, Wang Y, Titmus M, Botchkina G, Formentini A, Kornmann M, Ju J. Molecular mechanism of chemoresistance by miR-215 in osteosarcoma and colon cancer cells. Mol. Cancer. 2010;9:96. doi: 10.1186/1476-4598-9-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mishra PJ, Humeniuk R, Longo-Sorbello GS, Banerjee D, Bertino JR. A miR-24 microRNA binding-site polymorphism in dihydrofolate reductase gene leads to methotrexate resistance. Proc. Natl. Acad. Sci. USA. 2007;104:13513–13518. doi: 10.1073/pnas.0706217104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mishra PJ, Humeniuk R, Mishra PJ, Longo-Sorbello GS, Banerjee D, Bertino JR. A miR-24 microRNA binding-site polymorphism in dihydrofolate reductase gene leads to methotrexate resistance. Proc. Natl. Acad. Sci. USA. 2007;104:13513–13518. doi: 10.1073/pnas.0706217104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mishra PJ, Song B, Wang Y, Humeniuk R, Banerjee D, Merlino G, Ju J, Bertino JR. MiR-24 tumor suppressor activity is regulated independent of p53 and through a target site polymorphism. PLoS One. 2009;4:e8445. doi: 10.1371/journal.pone.0008445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lal A, Navarro F, Maher CA, Maliszewski LE, Yan N, O'Day E, Chowdhury D, Dykxhoorn DM, Tsai P, Hofmann O, Becker KG, Gorospe M, Hide W, Lieberman J. miR-24 Inhibits cell proliferation by targeting E2F2, MYC, other cell-cycle genes via binding to "seedless" 3'UTR microRNA recognition elements. Mol. Cell. 2009;35:610–625. doi: 10.1016/j.molcel.2009.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Donnenberg VS, Donnenberg AD. Multiple drug resistance in cancer revisited: the cancer stem cell hypothesis. J. Clin. Pharmacol. 2005;45:872–877. doi: 10.1177/0091270005276905. [DOI] [PubMed] [Google Scholar]
- 46.Dalerba P, Dylla SJ, Park IK, Liu R, Wang X, Cho RW, Hoey T, Gurney A, Huang EH, Simeone DM, Shelton AA, Parmiani G, Castelli C, Clarke MF. Phenotypic characterization of human colorectal cancer stem cells. Proc. Natl. Acad. Sci. USA. 2007;104:10158–10163. doi: 10.1073/pnas.0703478104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ansell A, Farnebo L, Grenman R, Roberg K, Thunell LK. Polymorphism of FGFR4 in cancer development and sensitivity to cisplatin and radiation in head and neck cancer. Oral Oncol. 2009;45:23–29. doi: 10.1016/j.oraloncology.2008.03.007. [DOI] [PubMed] [Google Scholar]
- 48.Song B, Wang Y, Titmus MA, Botchkina G, Formentini A, Kornmann M, Ju J. Molecular mechanism of chemoresistance by miR-215 in osteosarcoma and colon cancer cells. Mol. Cancer. 2010;9:96. doi: 10.1186/1476-4598-9-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sansam CL, Shepard JL, Lai K, Ianari A, Danielian PS, Amsterdam A, Hopkins N, Lees JA. DTL/CDT2 is essential for both CDT1 regulation and the early G2/M checkpoint. Genes Dev. 2006;20:3117–3129. doi: 10.1101/gad.1482106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Song B, Wang Y, Xi Y, Kudo K, Bruheim S, Botchkina GI, Gavin E, Wan Y, Formentini A, Kornmann M, Fodstad O, Ju J. Mechanism of chemoresistance mediated by miR-140 in human osteosarcoma and colon cancer cells. Oncogene. 2009;28:4065–4074. doi: 10.1038/onc.2009.274. [DOI] [PMC free article] [PubMed] [Google Scholar]