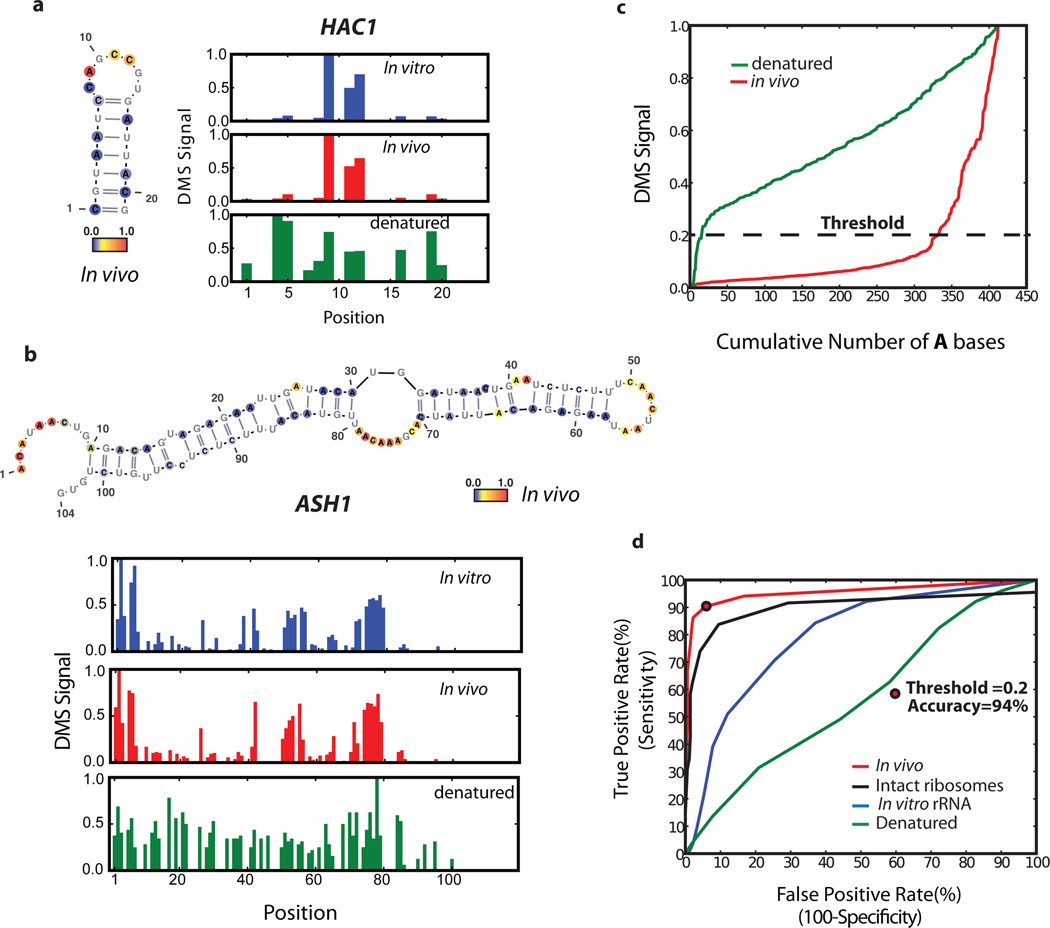
Figure 2. Comparison of DMS-seq data to known RNA structures.
A–b, DMS signal in (a) HAC1 (position 1 corresponds to chrVI:75828) (b) ASH1 (position 1 corresponds to chrXI:96245). Number of reads per position was normalized to the highest number of reads in the inspected region, which is set to 1.0. Also shown are the known secondary structures with nucleotides color-coded reflecting DMS-seq signal in vivo. c, DMS signal on 18S rRNA A bases plotted from least to most reactive. d, ROC curve on the DMS signal for A/C bases from the 18S rRNA. Threshold at 94% accuracy corresponds to 0.2 for the A bases.