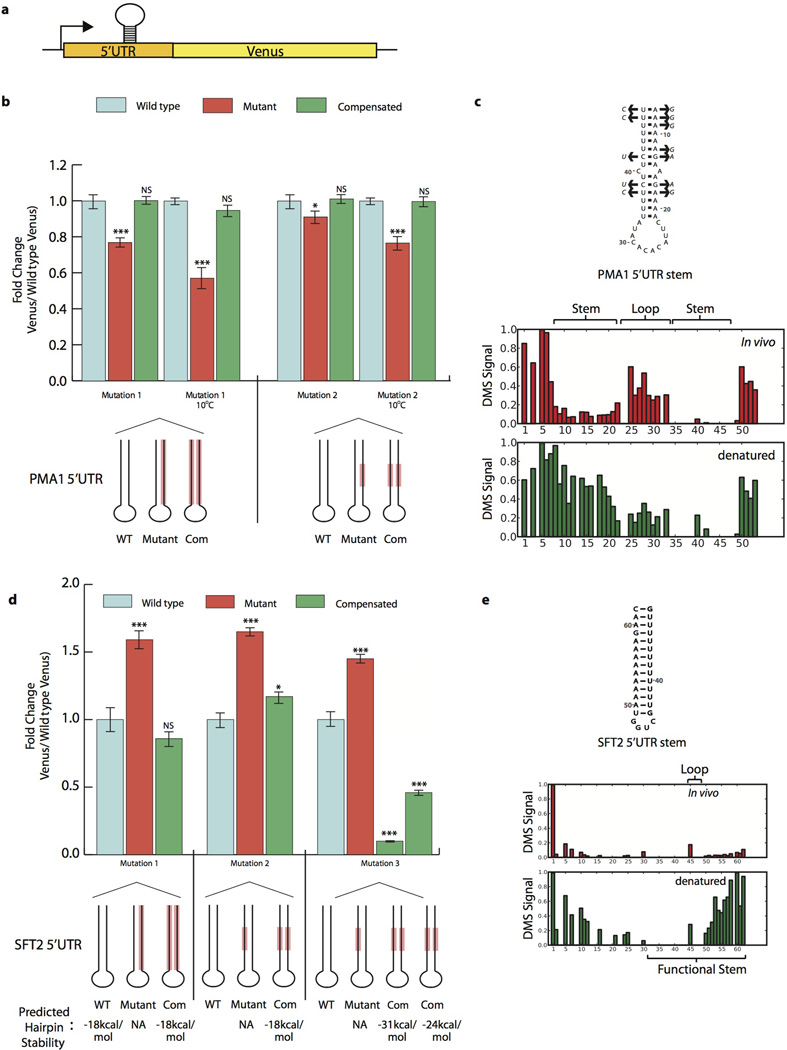
Extended Data Figure 5. Functional verification of novel 5’UTR structures in vivo.
a, Putative 5’UTR stems were manipulated in the context of a Venus reporter in vivo. b, PMA1 5’UTR structure was mutated and compensated twice with Venus reporter, differing in number and character of bases mutated. Mutation location shown in red on schematic. Reported p-values relative to wildtype Venus levels, calculated by two-sided t-test (p < .01, .001, and .0001 represent *, **, and *** respectively). For all graphs, Venus signal normalized to cell size before calculating fold change and data presented is from two biological and two technical replicates. Error bars represent SEM. c, Secondary structure of functional PMA1 5’UTR stem, with compensatory mutations (arrows) found in S. paradoxus, S. mikatae, S. kudriavzevii, and S. bayanus. Raw DMS signal shown below (position 1 = chrVII:482745). d, SFT2 5’UTR structure was mutated and compensated three times in Venus reporter system, differing in number, character, and location of bases mutated. Mutation location shown in red on schematic. Stem stability as predicated by mfold. Reported p-values relative to wild type Venus levels, also by two-sided t -test (p < .01, .001, and .0001 represent *, **, and *** respectively). Error bars represent standard deviation c, Secondary structure of functional SFT2 5’UTR stem. Position 1 = chrII:24023.