Fig. 4.
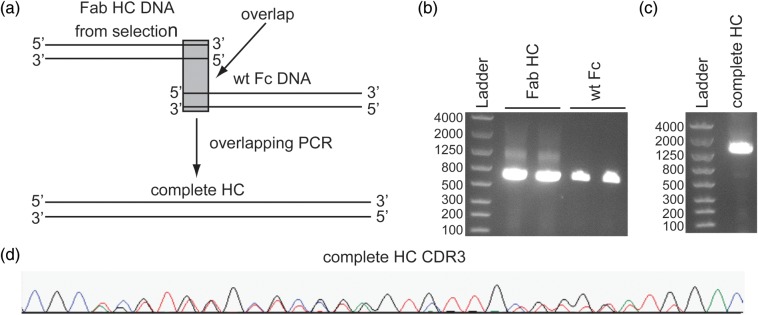
Reformatting selected Fab HCs to IgGs by overlapping PCR. (a) Overlapping PCR was used to generate the complete HC for subsequent IgG expression. (b) The Fab HC DNA was produced by RT-PCR of the output mRNA from Round 5 of the selection (expected size ∼670 bps). The wt Fc DNA including the hinge region and an overlapping segment of ∼30 bps into the CH1 domain was produced by PCR from a wt plasmid (expected size ∼700 bps). (c) The final assembled complete HC DNA is the main product and the correct size (expected size ∼1353 bps). (d) As desired, pooled sequencing of the assembled complete HC DNA does not indicate any significant differences from before assembly in the ratios of nucleotides observed throughout CDR3 (compared to Round 5, Fig. 3b).
