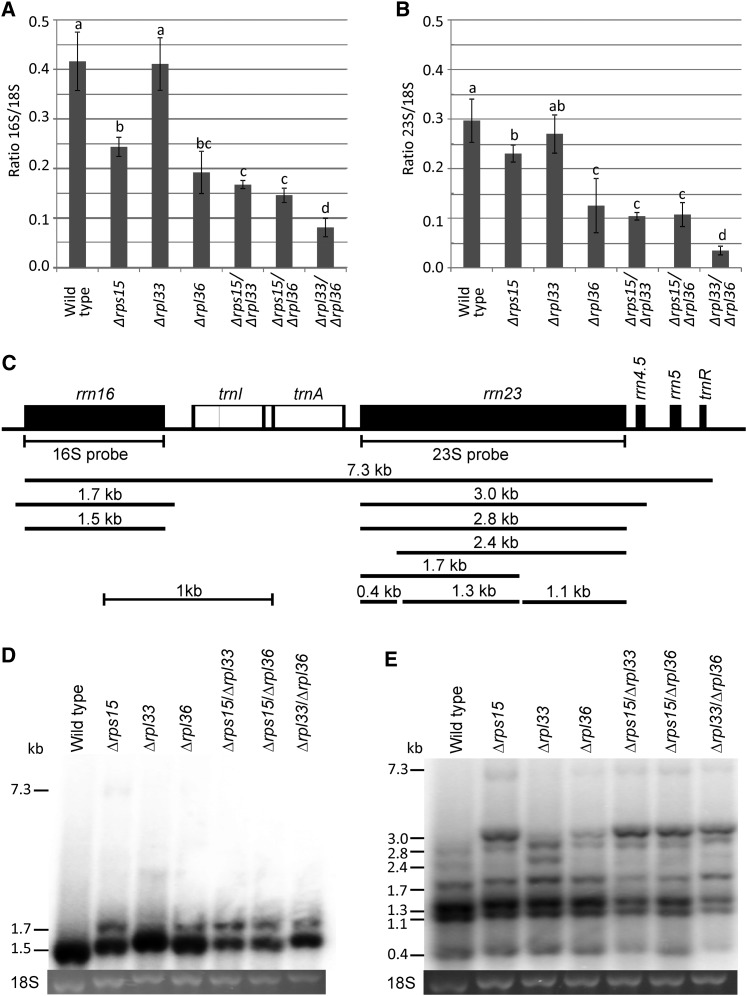
Figure 3.
Accumulation and Processing of rRNAs in Ribosomal Protein Mutants Grown Heterotrophically on Synthetic Medium.
Wild-type plants, single mutants of rps15, rpl33, and rpl36, and the three double mutants were analyzed.
(A) Accumulation of the 16S rRNA as a proxy for the accumulation of the 30S ribosomal subunit. The values give the ratio of the plastid 16S rRNA to the cytosolic 18S rRNA and represent the means of three biological replicates. The error bars indicate the sd. Statistically significant differences (determined by one-way ANOVA and Fisher’s LSD test, P < 0.05) are indicated by the letters above the bars.
(B) Accumulation of the 23S rRNA as a proxy for the abundance of 50S ribosomal subunits.
(C) Physical map and transcript processing pattern of the plastid rRNA operon. The 7.3-kb primary transcript, the different processing intermediates, and the mature forms of the 16S and 23S rRNAs are shown. The 23S rRNA is cut into three pieces, a phenomenon known as “hidden break” processing (Delp and Kössel, 1991). The positions of the hybridization probes for the 16S and 23S rRNAs are also indicated.
(D) Accumulation and processing of the plastid 16S rRNA determined by RNA gel blot hybridization. As loading control, the ethidium bromide–stained gel region containing the cytosolic 18S rRNA is shown.
(E) Analysis of the accumulation and processing pattern of the 23S rRNA. Note quantitative differences in the efficiency of hidden break processing, which is known to be influenced by developmental cues (Rosner et al., 1974).