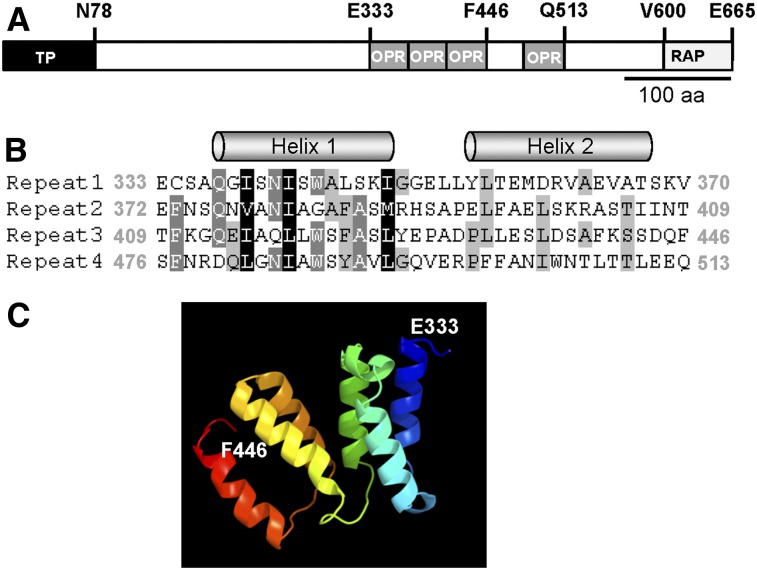
Figure 1.
Structural Features of the Arabidopsis RAP Protein.
(A) RAP protein structure. The plastid transit sequence predicted by TargetP (Emanuelsson et al., 1999) is shown as a black box. OPR repeats are depicted as gray boxes and the C-terminal RAP domain as a light-gray box. Several characteristic amino acids are indicated above the diagram. aa, amino acids.
(B) Sequence alignment of the four OPR repeats in RAP displayed with GeneDoc (Nicholas and Nicholas, 1997). Amino acid positions describing the beginning and end of the respective repeat are indicated in gray. The two α-helices in each repeat predicted by Jpred (www.compbio.dundee.ac.uk/www.jpred; Cole et al., 2008) are depicted above the sequences.
(C) 3D protein structure prediction. The structure model for the consecutive OPR tract including repeats 1 to 3 (amino acids 333 to 446) was predicted with the Phyre2 server (http://www.sbg.bio.ic.ac.uk/phyre2/; Kelley and Sternberg, 2009). The first (E333) and last (F446) amino acids are indicated. Rainbow coloring is shaded from blue (N-terminal site) to red (C-terminal site).