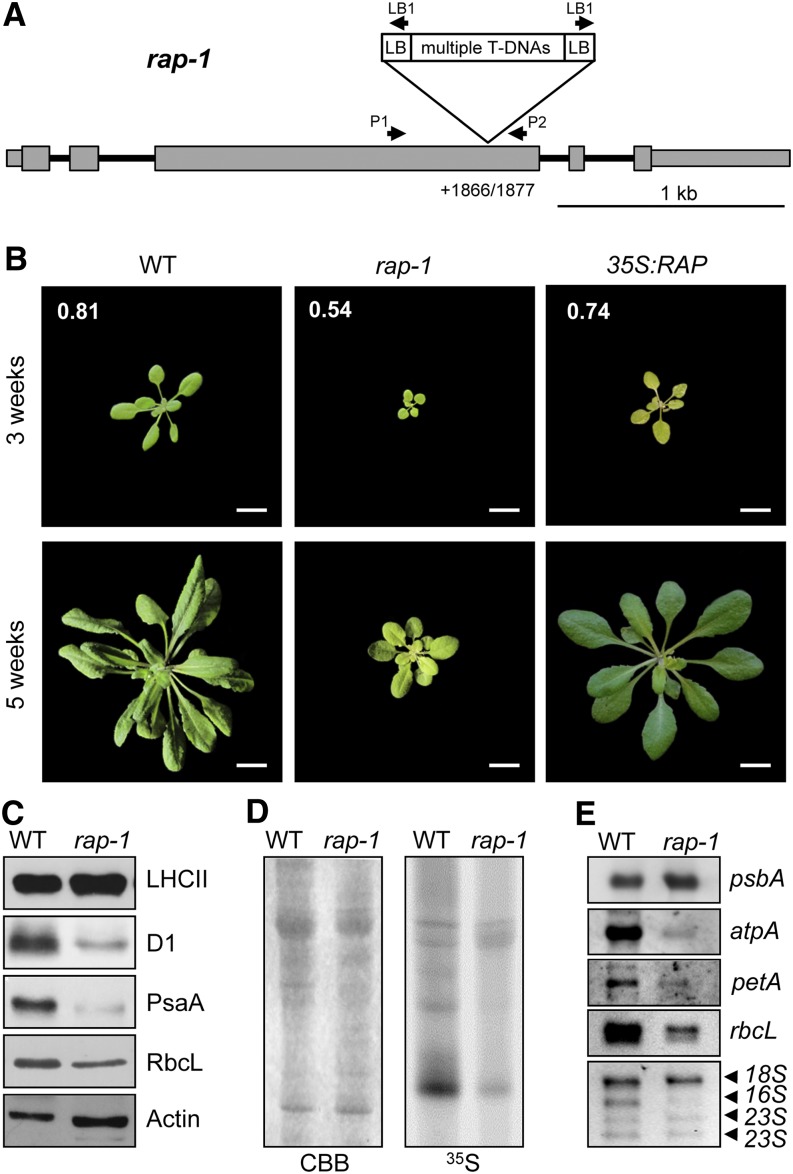
Figure 2.
Characterization of the rap-1 Mutant.
(A) Schematic depiction of the T-DNA insertion site in rap-1. Exons are shown as gray boxes, 5′ and 3′ UTRs as thinner gray boxes, and introns as black lines. The exact position of the T-DNA insertion site within the RAP gene in rap-1, identified by sequencing the DNA flanking the insertion site (Supplemental Figure 2A), is indicated (position +1866/1877 with respect to the translation initiation site). Primers used to identify homozygous mutants are indicated by the arrows above the gene model (Supplemental Figure 2B). The T-DNA insert is not drawn to scale.
(B) Growth phenotype of rap-1 and its complementation. The wild type, the rap-1 mutant, and rap-1 complemented with RAP cDNA (35S:RAP) were grown for 3 and 5 weeks as indicated. Fv/Fm values for 3-week-old plants are shown in white numbers on the top three photographs. Bars = 1 cm.
(C) Accumulation of chloroplast-encoded proteins in rap-1. Total protein extracts (30 µg) from 3-week-old wild-type and rap-1 plants were subjected to immunoblot analysis using antibodies against the proteins indicated on the right. β-Actin was used as the loading control. The RbcL protein was detected with an antiserum raised against the spinach Rubisco holoenzyme on a parallel, identical blot.
(D) In vivo translation assay. 35S-labeled thylakoid proteins from wild-type and rap-1 plants were separated by SDS-PAGE. The Coomassie blue–stained gel (CBB) and the autoradiograph (35S) are shown.
(E) Accumulation of chloroplast transcripts in rap-1. Total RNAs from 3-week-old wild-type and rap-1 plants were subjected to RNA gel blot analysis using the gene-specific probes indicated on the right. rRNAs on the ethidium bromide–stained gel were used as loading control (bottom panel).