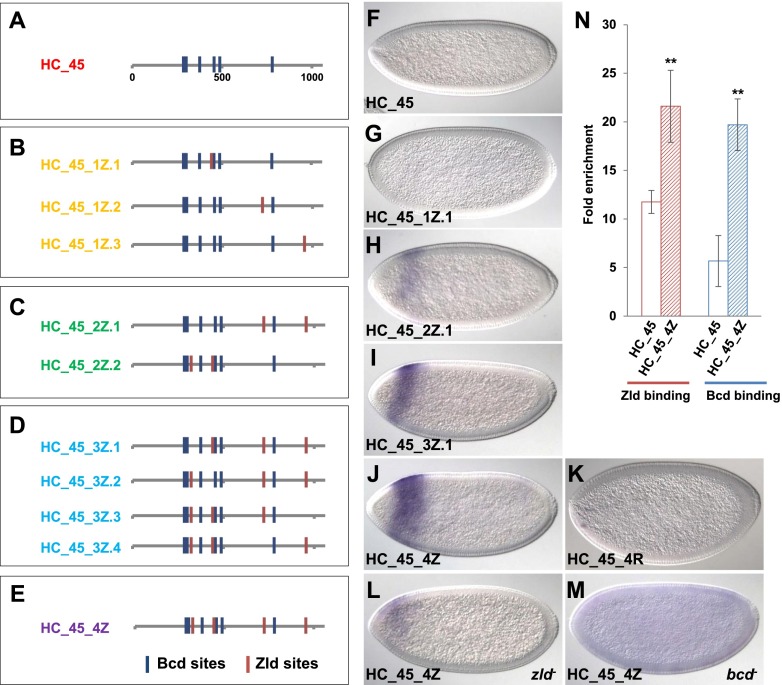
Figure 2.
Adding Zld sites activates the inactive fragment HC_45 and fine-tunes enhancer activity. (A–E) Maps of HC_45 variants that carry different numbers and combinations of extra Zld sites. Blue bars represent Bcd-binding sites (P < 0.005), and red bars represent Zld sites introduced to the HC_45 element. The number of extra Zld sites increases from 0 to 4, corresponding to A–E. Within each box, different combinations of Zld sites are shown. (F–J) lacZ RNA expression in a wild-type embryo carrying HC_45-lacZ transgenes with increasing numbers of additional Zld sites. (K) lacZ expression driven by HC_45 with scrambled sequences in the nucleotides that were mutated to add new Zld sites in J. (L,M) lacZ expression driven by HC_45_4Z in zld− (L) and bcd− (M) embryos. Note that there is still some expression of the HC_45_4Z construct in zld mutants. This suggests that another unknown factor can bind the inserted sites. (N) Zld (in red) and Bcd (in blue) ChIP-qPCR with HC_45 (blank bars) and HC_45_4Z (hatched bars). Primers were chosen to amplify only the transgenic fragment. A flanking region near the landing site was used as negative control (see the Supplemental Material for sequences). Zld- and Bcd-binding signals were normalized with the negative control and expressed as fold enrichment (see the Materials and Methods). Error bars represent standard deviations. Significant differences (P < 0.05, Student's t-test) are shown by double asterisks.