Figure 1.
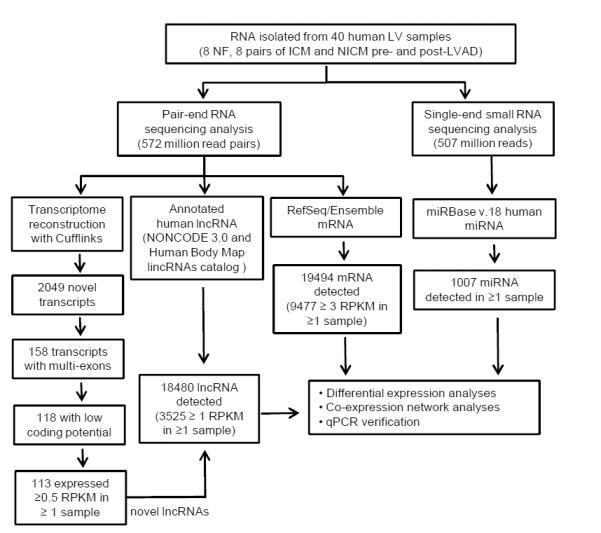
Workflow of comprehensive quantification of miRNAs, mRNAs and lncRNAs in human LV. Total RNA was isolated from non-failing (n=8) LV, and from ICM (n=8) and NICM (n=8) LV, before and after LVAD support; the latter were matched samples from the same patient. Poly-A(+) RNA libraries were constructed for pair-end RNA sequencing, whereas small RNA libraries were prepared for single-end sequencing. Pair-end sequencing reads went through transcriptome reconstruction and computational prediction pipelines to identify novel transcripts; among the 2049 novel transcripts uncovered by RNASeq, 113 were identified as novel lncRNAs. A compiled lncRNA annotation database, including NONCODE3.0, the Human Body Map lincRNAs catalog and the 113 novel lncRNAs identified in this study, was generated and utilized for quantitatve analyses of lncRNAs. RefSeq and the Ensemble database were used for quantification of mRNAs, and miRBase v.18 was used for miRNA quantification analyses. Results from RNASeq were subjected to differential expression and co-expression network analyses. In addition, quantitative PCR analysis of selected transcripts was conducted and compared with the results from RNASeq.
