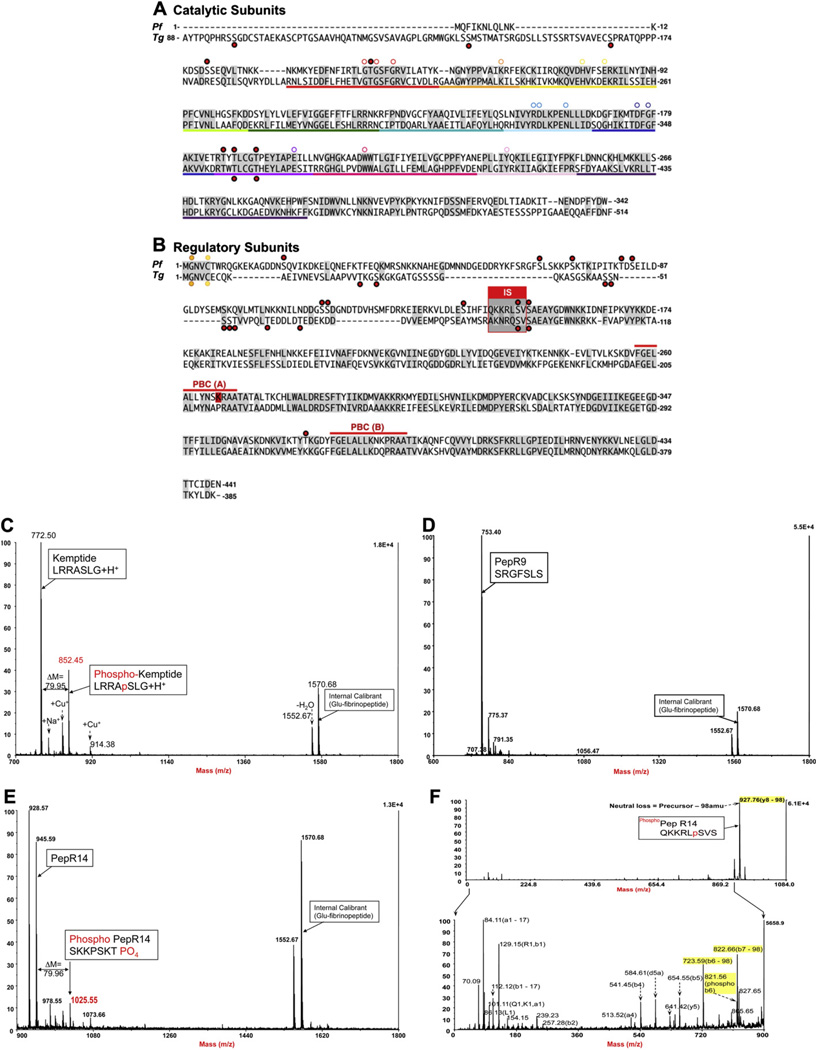
Fig. 6.
Putative phosphorylation sites of the catalytic and regulatory subunits of PfPKA and TgPKA, as detected by Treeck et al, 2011. (A) Sequences of PfPKA-C and TgPKA-C and (B) PfPKA-R and TgPKA-R with key conserved residues highlighted by a white circle and phosphorylation sites indicated by red dots. S149 in PfPKA-R is a bona fide PKA substrate. Spectra of PfPKA-R peptides, PepR149 (QKKRLSVS) harboring S149, PepR68 (SRGFSLS) harboring S68 and Kemptide (LRRASLG) following in vitro phosphorylation by bovine PKA. (C) MS trace of Kemptide used as positive control. The presented spectrum is a composite of two distinct spectra, since the phosho Kemptide (m/z = 852.5) was eluted around 1 min before its non-phosphorylated counterpart (m/z = 772.5). (D) MS trace of pepR68 used as negative control. PepR68 was visible in MS on a few consecutive fractions at around 28min elution. The presented spectrum shows the non-phosphorylated pepR68 (m/z = 753.4), while the phosphorylated form was not detected in any of the fractions. (E) MS trace of pepR149. Non-phosphorylated PepR149 was visible in MS on a few consecutive fractions at 20min elution. The presented spectrum is an intermediary fraction between the two elution peaks. Phosphorylated PepR149 (m/z = 1025.55) was eluted 30 s before its non-phosphorylated counterpart (m/z = 945.59). (D) MS/MS trace of CID-fragmented phosphoPepR149 (m/z = 1025.55); (F1) full spectrum showing an intense signal corresponding to H2PO4 (98amu) loss; (F2) zoomed spectrum below neutral loss m/z showing fragmented ions’ assignation of b and y ions according to Roepstorff and Fohlman [62], nomenclature correlating to PepR14. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)