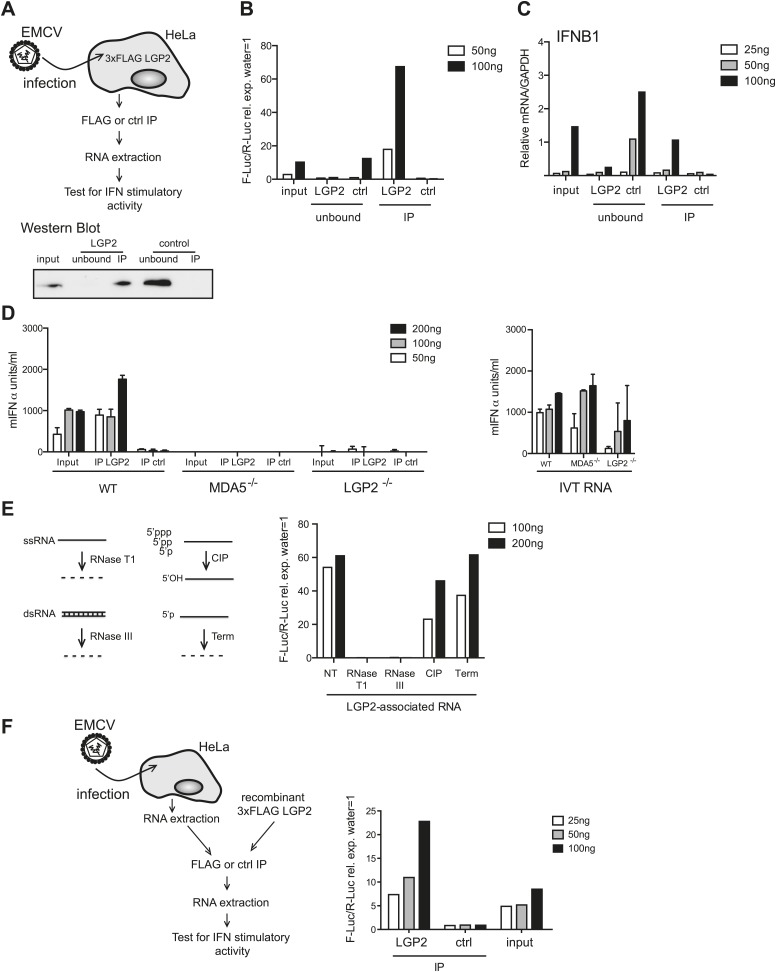
Figure 2. LGP2 pulldown captures MDA5 agonistic RNA from EMCV-infected cells.
(A) Schematic representation of the experimental setup for LGP2 immunoprecipitation (IP). Precipitation efficiency was routinely verified by immunoblotting with an anti-FLAG antibody; an example is shown in the lower panel. (B and C) The indicated amounts of RNA from EMCV-infected FLAG-LGP2-expressing HeLa cells (input), RNA associated with LGP2 or control (ctrl) immunoprecipitations (IP), or RNA remaining after LGP2 or control precipitations (unbound) were tested for the ability to stimulate the IFN-β promoter reporter assay in HEK293 cells (B) or induce IFNB1 mRNA (normalised to GAPDH) in HeLa cells (C). (D) The indicated amounts of RNA from samples processed as in (B) and (C) were transfected into WT, LGP2-deficient, or MDA5-deficient MEFs. Supernatants were harvested 16 hr later and mIFN-α levels measured by ELISA (left panel). IVT RNA transfections were used as positive controls (right panel). Error bars represent the standard deviation of three replicate transfections. (E) LGP2-associated RNA was not treated (NT) or digested with RNaseT1 (specific for ssRNA), RNaseIII (specific for base-paired RNA), CIP or Terminator (Term, an RNase specific for 5′monophosphate RNA) and subsequently tested for the ability to stimulate the IFN-β reporter in HEK293 cells. The activity of all enzymes was validated in control samples (not shown). (F) RNA from HeLa cells infected with EMCV (input) was incubated with recombinant FLAG-tagged LGP2 protein and anti-FLAG or control IP was performed as in (A). IP-associated RNA was isolated and tested, in parallel with input RNA, at the indicated doses in the IFN-β promoter luciferase reporter assay on HEK293 cells. Schematic representation of the experiment is shown on the left, results are presented on the right. One representative of the three (A–C) or two (D–F) experiments is shown.