Table 1.
Sub-circuit repertoire for developmental GRNs
| Regulatory state specification function |
Sub-circuits | What they do | Topologies | |
|---|---|---|---|---|
| X,1 − X processors | Double negative gate1,2,6 | Install regulatory state in X domain, prohibit same state everywhere else* | 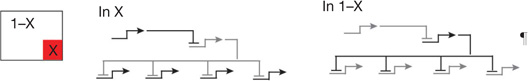 |
|
| 1.1 | ||||
| Signal-mediated switch2 | Activate regulatory gene(s) in cells receiving signal, repress same genes everywhere else† | 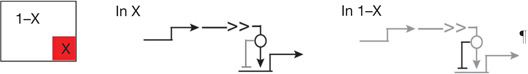 |
||
| 1.2 | ||||
| Spatial subdivision | Inductive signaling2 | Activation of new regulatory genes in a cellular domain by transcriptional response to signal ligands produced by other cells | 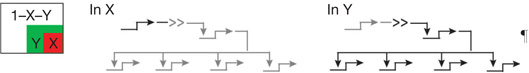 |
|
| 2.1 | ||||
| AND logic circuitry2 | Overlapping but spatially non-coincidental inputs are generated and both are required for regulatory gene activation, which occurs only in overlap subdomain | 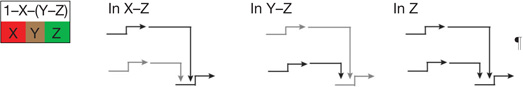 |
||
| 2.2 | ||||
| Spatial repression2 | Boundaries of spatial regulatory state domains controlled by transcriptional repression. | 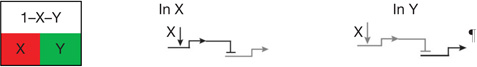 |
||
| 2.3 | ||||
| Dynamic lockdown of regulatory state‡ | Reciprocal repression of state1,2,9,39,51 | In each spatial regulatory state domain key activators of alternative states are transcriptionally repressed by ‘exclusion’ circuitry§. | 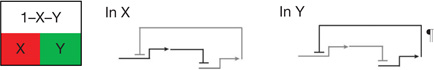 |
|
| 3.1 | ||||
| Feedback circuitry1 | Two or three regulatory genes engage in positive intergenic feedback, stabilizing regulatory state irrespective of transient inputs | 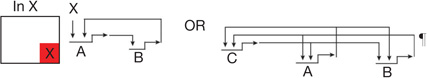 |
||
| 3.2 | ||||
| Community effect circuitry2,23,92 | Cells within a territory all signal to one another, driving continued uniform expression both of ligand gene and signal-dependent regulatory genes | | | 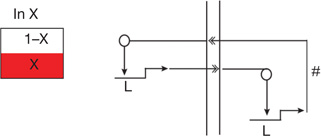 |
||
| 3.3 | ||||
| Boundary maintenance | Reciprocal repressive signalling across boundary32 | Different signals are produced by apposing cells and their reception triggers repressive circuitry excluding the cross-boundary regulatory state | 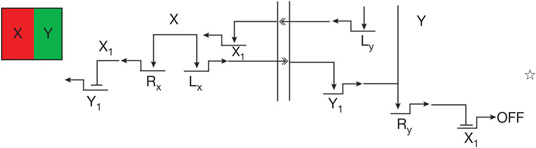 |
|
| 4 | ||||
| Terminal binary cell fate choice | Alternate sub-circuits driven by reciprocal repressors8,36–38,81 | External inputs tip the balance of repressor expression, resulting in activation of one differentiation program and exclusion of the other | 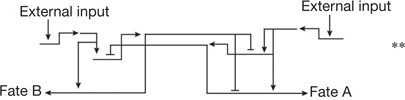 |
|
| 5 | ||||
| Discontinuous transcriptional response to signal intensity and/or duration | Reciprocal repressor genes responding cooperatively to inducer21,38,81 | Circuitry generates differential stimulation of expression of reciprocal repressors in low versus high signal intensity†† | 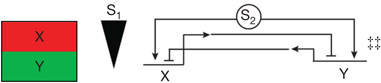 |
|
| 6.1 | ||||
| Reciprocal repressor genes, one activating an additional repressor gene, each with variable external positive inputs82 | Circuitry generates irreversible transitions, in stem cell regulatory state, off versus on in response to signals of different strength and duration | 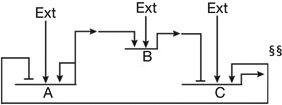 |
||
| 6.2 | ||||
| Triple feedback linkage with asymmetric signal inputs47 | Produces alternative regulatory states, or low level indeterminate state, depending of different positive inputs | | | | | 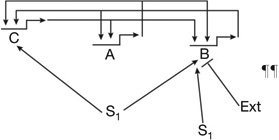 |
||
| 6.3 | ||||
The role of the sub-circuit is given in column 1; its name in column 2; a description of its function in column 3; and the sub-circuit structure in column 4. Numbers in column 2 are keyed to Fig. 1. See references indicated for actual occurrences, exact circuit topologies, and discussion of information processing specifics. In each case the functions of the circuit are hardwired in its cis-regulatory target sites. In Topologies column, all genes encode transcription factors unless otherwise noted.
Regulatory genes that create initial regulatory state are controlled by widely expressed repressor, which is dominant over their positive inputs, and gene encoding this repressor is itself specifically repressed in a local region (X) by another gene encoding a different repressor: hence target genes are ON in X, specifically repressed elsewhere.
Many developmental signalling systems (for example, Notch, Wnt) activate immediate early response factors in cells receiving ligand, but in absence of ligand, these factors act as dominant repressors of the same target genes.
Dynamic in that continuing transcription is required.
Exclusion sub-circuits are activated as downstream outputs of specification GRNs.
A unique circuit design here is that the ligand gene is activated by the same signal transduction mechanism reception of the ligand activates in recipient cells; a positive intercellular feedback.
From ref 2.
L, gene encoding signalling ligand.
R encodes repressor; L encodes signalling ligand.
This example was adapted from Ref 36.
Conceived as a means of obtaining different discrete transcriptional responses from a graded signal; see discussion of this type of circuitry in section on mathematical models below.
S, signal; triangle represents graded signal strength
S1, S2, different signal inputs gene B is subject to additional transcriptional repression in certain regulatory states.
This design precludes necessity for ad hoc Hill coefficients as in 5, 6.1; see section on mathematical models below.
Autoregulatory loops lock on whichever state the system goes to.
