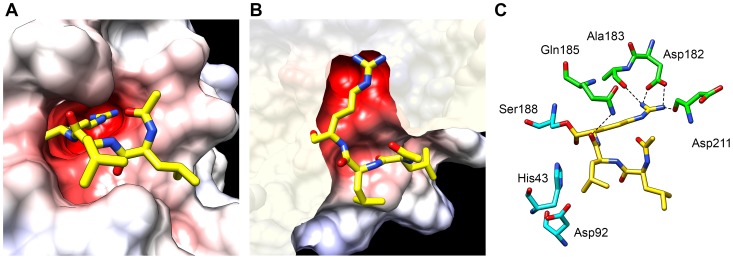
Figure 7. Homology model of the SmSP1 protease domain in complex with leupeptin.
The model was built using the template X-ray structure of bovine trypsin in complex with substrate-like inhibitor leupeptin (N-acetyl-L-leucyl-L-leucyl-L-argininal; PDB code 1jrt). (A) Surface representation of the SmSP1 active site colored by electrostatic potential (at a scale from −10 kT/e (red) to +10 kT/e (blue)). Carbon atoms of leupeptin are yellow; heteroatoms have a standard color coding (O, red; N, blue). (B) The same detail as (A) but viewed from above (the surface display was clipped for a better view). (C) Schematic view of the active site residues of SmSP1 (green) forming hydrogen bonds (dashed lines) with leupeptin (yellow). Note the interactions between Asp182 (in the S1 protease subsite) and the basic P1 residue of leupeptin that mimic the S1-P1 salt bridge that is critical for trypsin-like substrate specificity. Catalytic residues (cyan) are shown, including the covalent linkage of leupeptin with the catalytic Ser188.