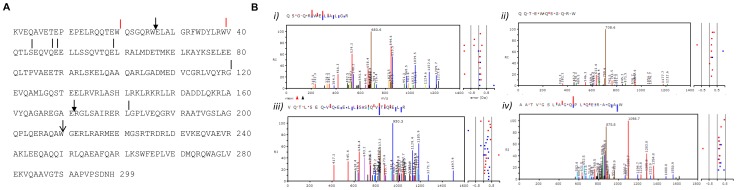
Figure 4. Detection of chymotrypsin cleavage sites in apoE3 using mass spectrometry.
A) Representation of full length apoE3 protein sequence and observed proteolytic sites in ∼25, 20 and 17 kDa fragments generated upon incubation of rE3 in hippocampal conditioned medium. Two common cleavages sites at the carboxy side of W-20 and W-39 are indicated by red lines. Additional cleavage site in the ∼25 kDa apoE fragments at W-210 is indicated by arrow. Additional 6 cleavage sites in ∼20 kDa fragments are indicated by black lines, whereas additional two cleavage sites at W-26 and E-171 in ∼17 kDa fragments are marked by arrows with solid heads. Amino acid position is indicated by numbers. B) Annotated tandem mass spectra of identified peptides generated upon cleavage by chymotrypsin like protease at W-20 (i), W-26 (ii) W-39 (iii) and W-210 (iv).