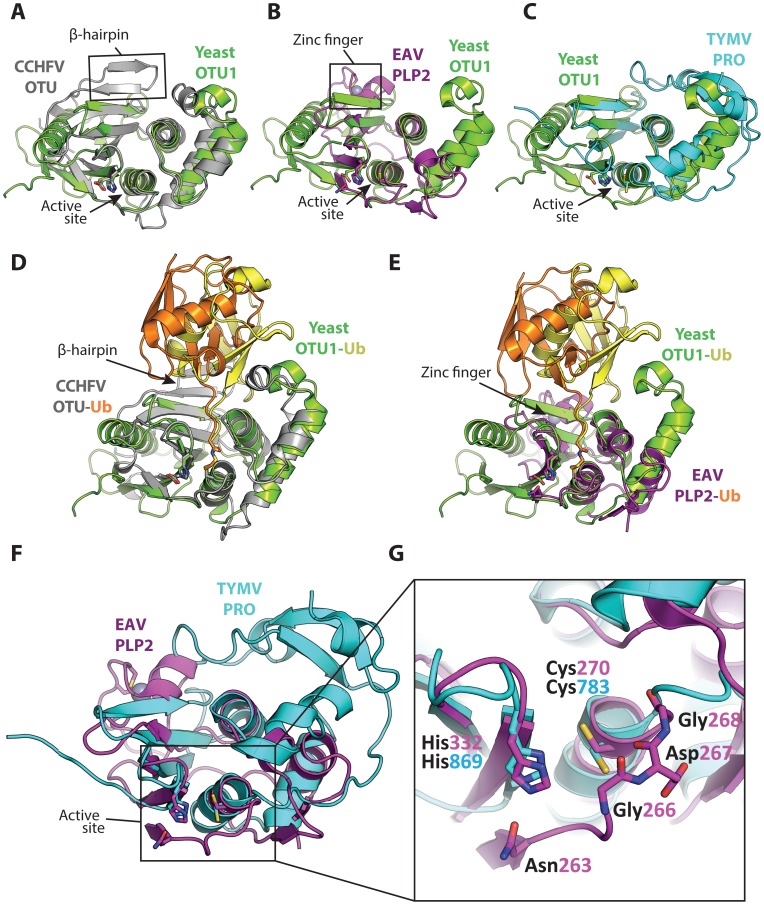
Figure 2. Superpositions of the viral OTU proteases with yeast OTU1 and one another.
Superpositions of yeast OTU1 (3BY4) [57] with (A) CCHFV OTU (3PT2) [23], RMSD: 1.8 Å over 112 residues, (B) EAV PLP2 (4IUM) [26], RMSD: 2.8 Å over 69 residues, and (C) TYMV PRO (4A5U) [27], [28], RMSD: 1.4 Å over 76 residues. Superpositions of the yeast OTU1-Ub complex with (D) the CCHFV OTU-Ub complex and (E) the EAV PLP2-Ub complex, highlighting the difference in the orientation of Ub between the two viral OTU domains versus the eukaryotic yeast OTU1 domain. The Ub that is complexed with yeast OTU1 is depicted in yellow, while the Ub complexed with CCHFV OTU or EAV PLP2 is depicted in orange. (F) Superposition of EAV PLP2 and TYMV PRO, RMSD: 2.5 Å over 53 residues. (G) Close-up of the active site region (boxed) of the superposition depicted in F. Side chains of the catalytic cysteine (Cys270 and Cys783 for EAV PLP2 and TYMV PRO, respectively) and histidine (His332 and His869 for EAV PLP2 and TYMV PRO, respectively) residues are shown as sticks, as well as the active site Asn263 for EAV PLP2. The backbone amide group of Asp267 likely contributes to the formation of the oxyanion hole in the active site of EAV PLP2, yet a functionally equivalent residue is absent in TYMV PRO. The Gly266 and Gly268 residues flanking Asp267 in EAV PLP2 are depicted as sticks as well, for clarity. Note the alternative orientation of the active site cysteine residue of TYMV PRO which, unlike EAV PLP2, was not determined in covalent complex with an Ub suicide substrate. All alignments were generated using the PDBeFOLD server [64], and thus the reported RMSD values differ from those reported in Table 1, in which the DALI server was used. The yeast OTU1, CCHFV OTU, and EAV PLP2 domains were all crystallized in complex with Ub, which has been removed in panels A, B, C, F, and G for clarity. All images were generated using PyMol [60]. RMSD, root-mean-square deviation.