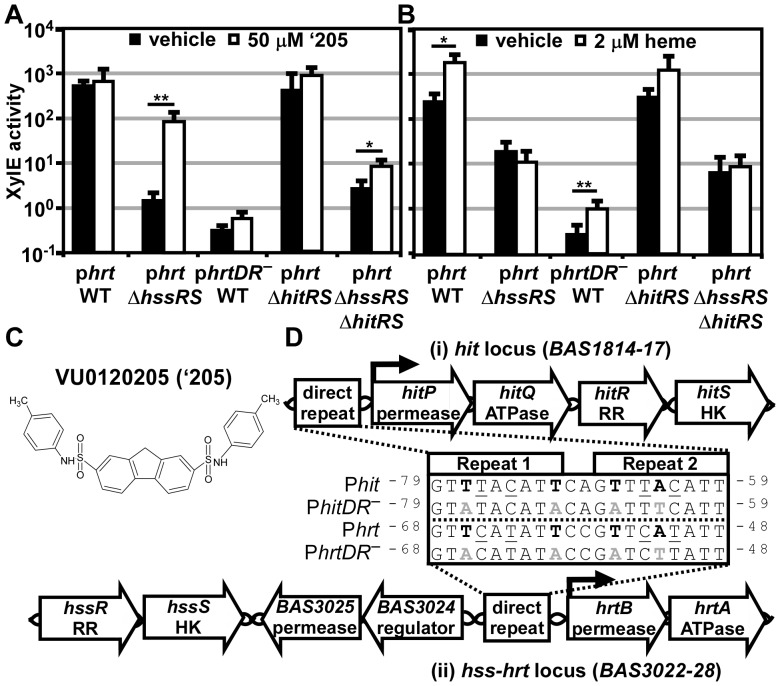
Figure 2. ‘205 activates the hrt promoter through HitRS in a manner that requires the direct repeat.
(A-B) The activity of Phrt in response to either 50 μM ‘205 (A) or 2 μM heme (B) was quantified. B.anthracis Sterne (WT), ΔhssRS, ΔhitRS, ΔhssRSΔhitRS were transformed with the xylE reporter plasmid, phrt. Four conserved nucleotides in the Phrt DR were mutated in the xylE reporter plasmid (phrtDR–) and transformed into WT (bold in D, inset). Each strain was grown in triplicate and exposed to the listed compound for 24 h and XylE activity was quantified. Error bars represent ±SD of data averaged from at least two independent experiments performed with biological triplicates. Significance was calculated by an unpaired Student's t-test, where * = P≤0.01 and ** = P≤0.001. (C) Depicted is the structure of VU0120205 (‘205). (D) Genomic loci of (i) hitPQRS (formerly Sterne BAS1814-17) and (ii) hssRS-hrtAB. The inset depicts an alignment of the DRs found in B. anthracis Phrt and Phit regions. Highlighted in bold are nucleotides previously shown to be required for HssR recognition of the Phrt DR [29]. The DR nucleotide mutations used in A and B are in gray, bold (DR–). Underlined are nucleotides that differ between Phit and Phrt. Superscript numbers indicate the nucleotide distance from the start site.