Figure 5. Overview of various optical tweezers instruments and their possible applications to studying translational elongation.
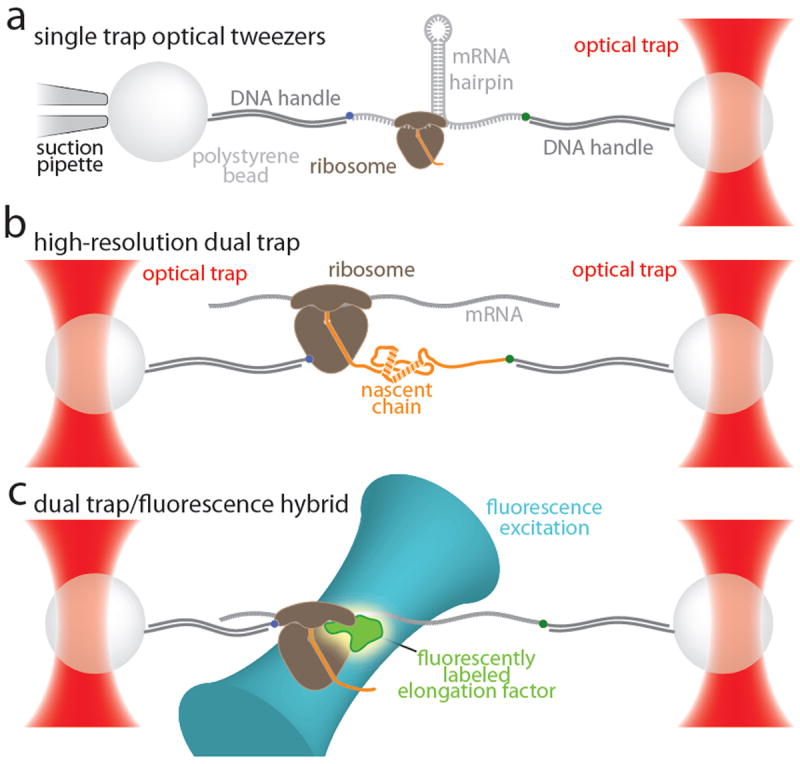
a. Single trap design. The sample is immobilized between two beads. One is held in an optical trap, the other is immobilized on a micro-pipette by suction. In the example shown here, an RNA hairpin is translated by the ribosome. Translocation of the ribosome by one codon results in opening of three base pairs of the hairpin, increasing the length of the RNA by six nucleotides, providing two-fold signal amplification that is intrinsic to this geometry. This geometry has successfully been used to study translation.5,99 Mechanical coupling of the experiment to the environment through the micro-pipette limits the spatial resolution and, perhaps more significantly, the drift stability of this geometry. b. Dual trap design. Both beads are held by optical traps. The traps can be formed by splitting a single laser beam, limiting the effects of fluctuations in laser power. In addition, the experiment is mechanically decoupled from the environment, resulting in superior drift stability. An ultra-stable instrument based on this design 112 can possibly be used to follow elongation of the nascent protein as depicted in the geometry shown here. The resulting signal would be a convolution of nascent chain elongation and folding. b. Hybrid instrument combining mechanical manipulation with single-molecule fluorescence detection. The design for a dual trap instrument that is capable of detecting the fluorescence of a single immobilized molecule between the two beads has recently been published.77 In the geometry suggested in this panel, the ribosome and the 3’-end of the mRNA are attached to beads. Translocation is observed as a stepwise decrease of the molecular extension; binding of exogenously labeled accessory factors is detected by fluorescence emission. This instrument may enable simultaneous measurements of translocation and binding of elongation factors and tRNAs.
