Figure 8. Modulation of nascent protein folding by the ribosome.
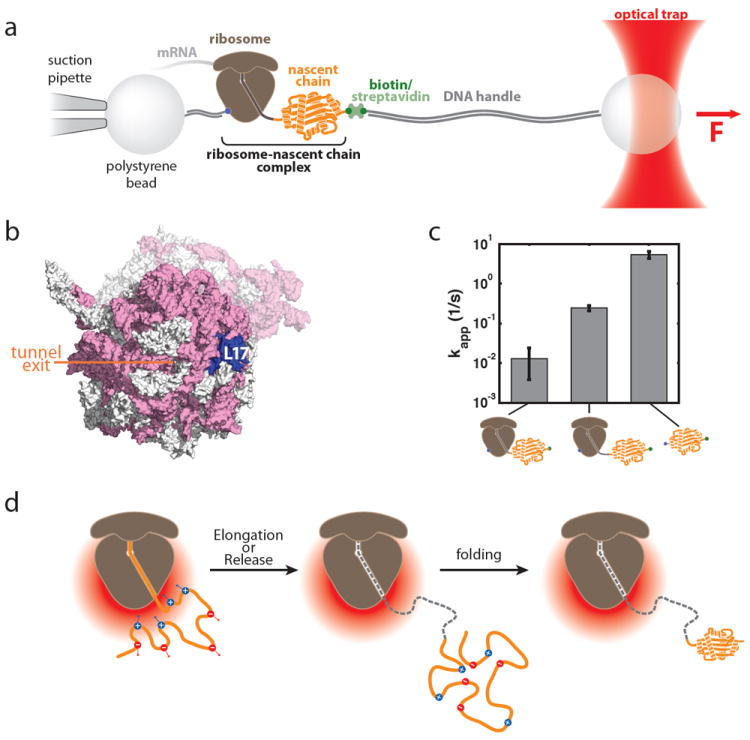
a. Optical tweezers geometry for measuring nascent chain folding. A single stalled ribosome-nascent chain complex is immobilized between polystyrene beads in a single-trap optical tweezers instrument. b. View of the area around the ribosomal exit tunnel. rRNA is shown in pink, ribosomal proteins in grey in this surface representation of the 70S ribosome (based on pdb coordinates 2aw4 and 2avy; ref. 107). L17, which was chosen as the attachment point on the ribosome, is shown in blue, the opening of the ribosomal exit tunnel is indicated. c. Apparent folding rate of T4 lysozyme bound to the ribosome (by linkers of 41 and 60 amino acids in length) and in the absence of the ribosome (from left to right). The ribosome reduces the apparent folding rate in a distance dependent fashion (see Reference 67 for details). d. Possible model for modulation of folding rates in an electrostatic potential. The negative electrostatic potential around the ribosome (red gradient) attracts positively charged residues within the nascent polypeptide and repels negatively charged ones. This leads to a structural bias that might affect the conformational search, which could make it energetically more difficult (or easier) to reach the transition state and thus result in a deceleration (or acceleration) of folding rates. Once the folding unit leaves the immediate environment around the ribosome (middle panel), the effect decays and folding proceeds in an unimpaired fashion (right panel). A, B and C: modified from Reference 67 (Adapted with permission from ref. 67. Copyright 2011 American Association for the Advancement of Sciences)
