Figure 2.
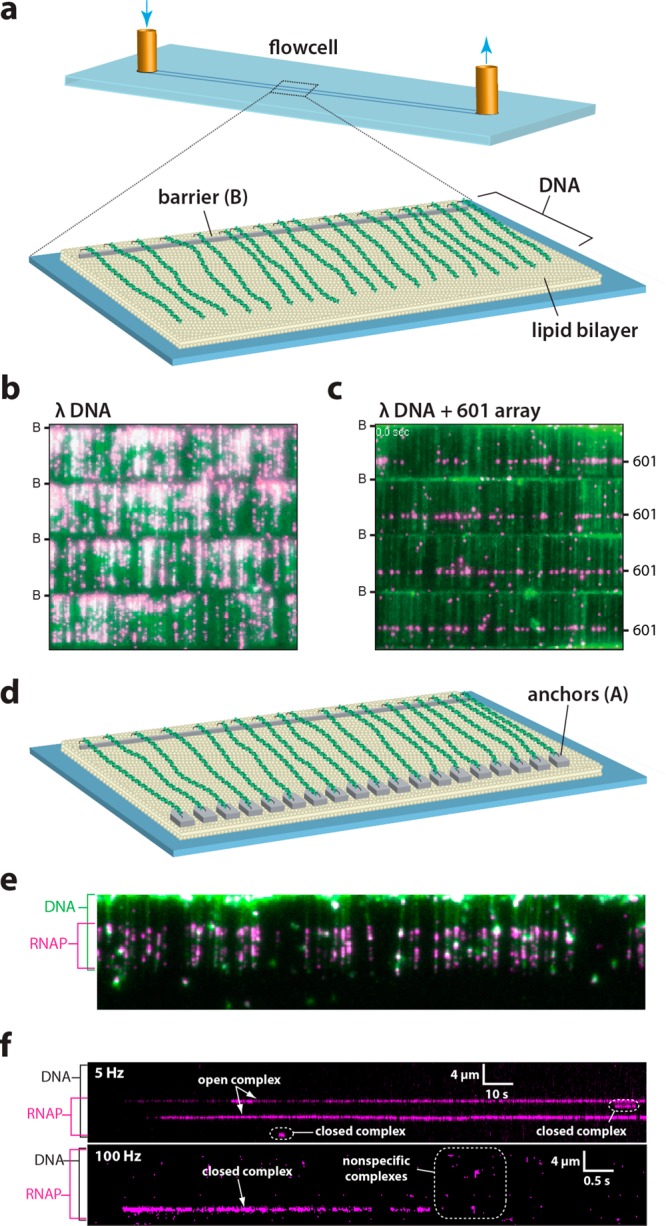
DNA curtains as a tool for studying protein–DNA interactions. (a) Schematic depiction a single-tethered DNA curtain in which one end of the DNA is anchored to a lipid bilayer and aligned along the leading edge of a nanofabricated barrier on the surface of the flow cell.37g (b) Example of a 4-tiered DNA curtain bound by recombinant nucleosomes each of which is tagged with a fluorescent quantum dot. Lambda phage DNA is stained with YOYO1 and shown in green, the nucleosomes are in magenta, and the location of the nanofabricated barriers (B) are indicated. Adapted with permission from ref (37i). (c) Example of a DNA curtain made using a lambda phage DNA substrate containing a 13× array of the Widom 601 nucleosome positioning sequence (unpublished). (d) Schematic depiction a double-tethered DNA curtain in which one end of the DNA is anchored to a lipid bilayer and aligned along the leading edge of a nanofabricated barrier and the second end of the DNA is anchored to an antibody-coated pedestal.37b (e) Wide-field TIRFM image showing quantum dot-tagged RNA polymerase (magenta) stably bound to the native promoters within the lambda phage genome. (f) Kymographs showing the association of RNA polymerase with an individual molecule of DNA (unlabeled) in real time. Adapted with permission from ref (37j).
