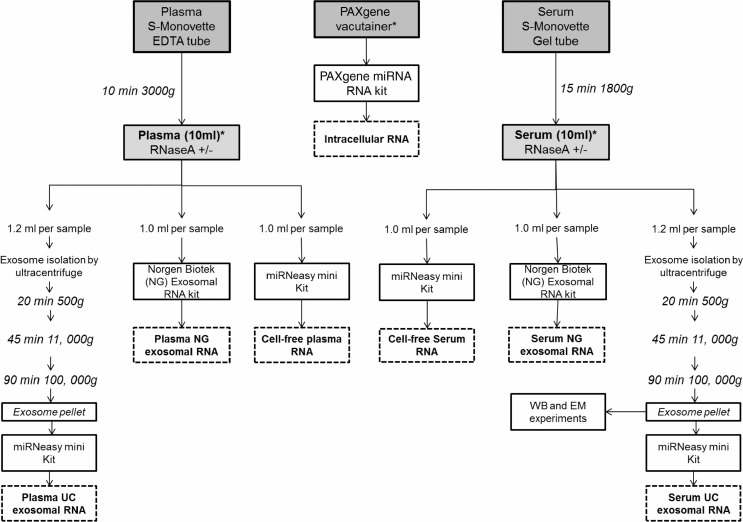
Fig. 1.
Work flow of study design and sample processing. Whole blood from 3 different individuals was collected by venepuncture into each tube using a Multi-fly and processed to analyse intracellular, cell-free and exosomal miRNA. Asterisks indicate the point of RNaseA treatment (100 ng/ml, 37°C for 10 minutes) to investigate RNA degradation in these samples. The workflow outlines the sample collection and preparation from 1 individual. The number of tubes collected from each volunteer was: 2×PAXgene 2.5 ml tubes, 3×Sarstedt S-Monovette serum-gel 7.5 ml tubes and 3×Sarstedt S-Monovette EDTA 7.5 ml tubes. Upon centrifugation of the Sarstedt S-Monovette EDTA tubes, approximately 10 ml of plasma was obtained across 3 Sarstedt S-Monovette tubes which are then separately aliquoted into Lo-Bind DNA tubes (4×1 ml, 2×1.2 ml tubes) for RNA analysis and deep sequencing. The remaining plasma was aliquoted for Western immunoblotting (WB, 1.2 ml), transmission electron microscopy (EM, 1.2 ml) and qNano (1 ml) analysis. For the RNA work involving RNaseA treatment, samples were allocated for an untreated control and RNaseA treatment: 2×1.2 ml for the ultracentrifugation exosomal RNA isolation (Plasma UC), 2×1 ml for the Norgen Biotek exosomal RNA isolation (Plasma NG), and 2×1 ml aliquot was reserved for cell-free plasma RNA isolation. The collection process and sample allocation are repeated for serum collection. Exosomes isolated from serum via the ultracentrifuge are denoted as Serum UC. Exosomal RNA isolated by the Norgen Biotek Kit are denoted as Serum NG. As for the 2×PAXgene tubes, RNA is isolated from 2.5 ml of whole blood per tube and isolated as recommended by the manufacturers protocol. One tube was treated with RNaseA and one was left untreated.